Team:TU Delft/22 July 2010 content
From 2010.igem.org
(Difference between revisions)
Ravandervalk (Talk | contribs) (→Alkane degradation) |
Ravandervalk (Talk | contribs) (→Salt tolerance) |
||
| Line 121: | Line 121: | ||
Time to get cracking, so bbc1 and B0015 were restricted using the scheme shown below. | Time to get cracking, so bbc1 and B0015 were restricted using the scheme shown below. | ||
| + | |||
| + | {| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | ||
| + | |'''#''' | ||
| + | |'''Sample added''' | ||
| + | |'''Volume added (µl)''' | ||
| + | |- | ||
| + | |1 | ||
| + | |bbc1 | ||
| + | |3 | ||
| + | |- | ||
| + | | | ||
| + | |Ecro R1 | ||
| + | |1 | ||
| + | |- | ||
| + | | | ||
| + | |Spe1 | ||
| + | |1 | ||
| + | |- | ||
| + | | | ||
| + | |Buffer 2 (biolabs) | ||
| + | |2 | ||
| + | |- | ||
| + | | | ||
| + | |Water | ||
| + | |11 | ||
| + | |- | ||
| + | | | ||
| + | |BSA | ||
| + | |2 | ||
| + | |- | ||
| + | |2 | ||
| + | |B0015 | ||
| + | |3 | ||
| + | |- | ||
| + | | | ||
| + | |Ecro R1 | ||
| + | |1 | ||
| + | |- | ||
| + | | | ||
| + | |Xba1 | ||
| + | |1 | ||
| + | |- | ||
| + | | | ||
| + | |Buffer 2 (biolabs) | ||
| + | |2 | ||
| + | |- | ||
| + | | | ||
| + | |Water | ||
| + | |11 | ||
| + | |- | ||
| + | | | ||
| + | |BSA | ||
| + | |2 | ||
| + | |} | ||
Revision as of 09:15, 10 August 2010
Lab work
Alkane degradation
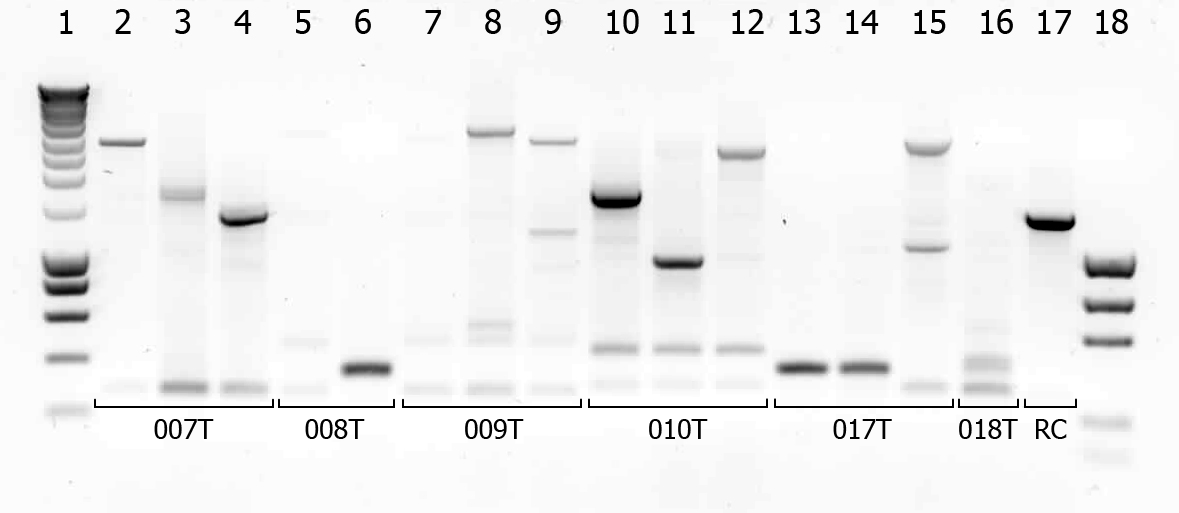
There were some colonies on Tuesday's plates! We had left the plates @ 37°C yesterday after having seen that there were no colonies. When checking this morning on all plates (except the negative control) there were a few colonies! (2-50 colonies). Chances are it's not what we're looking for, but maybe they are good transformants... to check we will do a colony PCR.
| # | Description | Expected length (bp) | Primers | Status |
| 1 | SmartLadder | n/a | n/a | n/a |
| 2 | Transformant #1 of ligation mix 007T | 1616 | G00100 + G00101 | |
| 3 | Transformant #2 of ligation mix 007T | 1616 | G00100 + G00101 | |
| 4 | Transformant #3 of ligation mix 007T | 1616 | G00100 + G00101 | |
| 5 | Transformant #1 of ligation mix 008T | 551 | G00100 + G00101 | |
| 6 | Transformant #2 of ligation mix 008T | 551 | G00100 + G00101 | |
| 7 | Transformant #1 of ligation mix 009T | 551 | G00100 + G00101 | |
| 8 | Transformant #1 of ligation mix 009T | 560 | G00100 + G00101 | |
| 9 | Transformant #1 of ligation mix 009T | 560 | G00100 + G00101 | |
| 10 | Transformant #1 of ligation mix 010T | 1657 | G00100 + G00101 | |
| 11 | Transformant #1 of ligation mix 010T | 1657 | G00100 + G00101 | |
| 12 | Transformant #1 of ligation mix 010T | 1657 | G00100 + G00101 | |
| 13 | Transformant #1 of ligation mix 017T | 1130 | G00100 + G00101 | |
| 14 | Transformant #1 of ligation mix 017T | 1130 | G00100 + G00101 | |
| 15 | Transformant #1 of ligation mix 017T | 1130 | G00100 + G00101 | |
| 16 | Transformant #1 of ligation mix 018T | 1874 | G00100 + G00101 | |
| 17 | Transformant #1 of Red colony | 1360 | G00100 + G00101 |
A number of colonies look promising! To check if they really are the BioBricks we want, tomorrow we will do a plasmid isolation with the cultures of lane 2, 4, 6, 7, 9 and 14. We will cut the isolated plasmids with various restriction enzymes and analyze the digestion products on gel.
Salt tolerance
Time to get cracking, so bbc1 and B0015 were restricted using the scheme shown below.
| # | Sample added | Volume added (µl) |
| 1 | bbc1 | 3 |
| Ecro R1 | 1 | |
| Spe1 | 1 | |
| Buffer 2 (biolabs) | 2 | |
| Water | 11 | |
| BSA | 2 | |
| 2 | B0015 | 3 |
| Ecro R1 | 1 | |
| Xba1 | 1 | |
| Buffer 2 (biolabs) | 2 | |
| Water | 11 | |
| BSA | 2 |
 "
"