|
JULY: WEEK 4
July, 19th
TECAN test showed that no RFP was produced from our parts, so all parts are potentially correct! For this reason we decided to sequence:
- I14-1 (Forward)
- I16-1 (Forward)
- I17-1 (Forward)
- I18-1 (Forward)
- I19-1 (Forward)
We also sequenced:
- I74C5-2 (Forward and Reverse)
- I84C5-2 (Forward and Reverse)
- I12-2 (Forward and Reverse)
These samples were prepared for sequencing (DNA was essicated) and sent to BMR genomics.
Trasformation of RING into:
- BW25141 (pir+)
- BW25142 (pir116)
- BW23474 (pir116)
- DH5alpha
- MG1655
Cultures were plated on:
- BW2514: Cm 34ug/ml
- BW25142: Cm 34ug/ml
- BW234741: Cm 34ug/ml
- DH5alpha: Cm 12,5ug/ml
- MG1655: Cm 12,5ug/ml
Inoculum of:
- I3-1
- I10-1
- I12-2
- I14-1
- I17-1
- <partinfo>BBa_J23110</partinfo>
in 5ml LB+Amp. Cultures were grown ON 37°C 220 rpm.
BW23473 arrived from Yale University on a paper disk. It was grown ON in 5ml L, at 37°C, 220 rpm.
July, 20th
 BW23474 transformed with <partinfo>BBa_J72007</partinfo> |  DH5alpha transformed with <partinfo>BBa_J72007</partinfo> |
Results for plates incubated ON, 37° C:
- BW23474 (pir116): showed colonies
- BW25141 (pir+): showed colonies
- BW25142 (pir116): showed colonies
- DH5alpha: didn't show colonies
- MG1655: didn't show colonies (even if there were a very few colonies that we suppose integrated the resistance of RING to survive - or the plate antibiotic wasn't homogeneous)
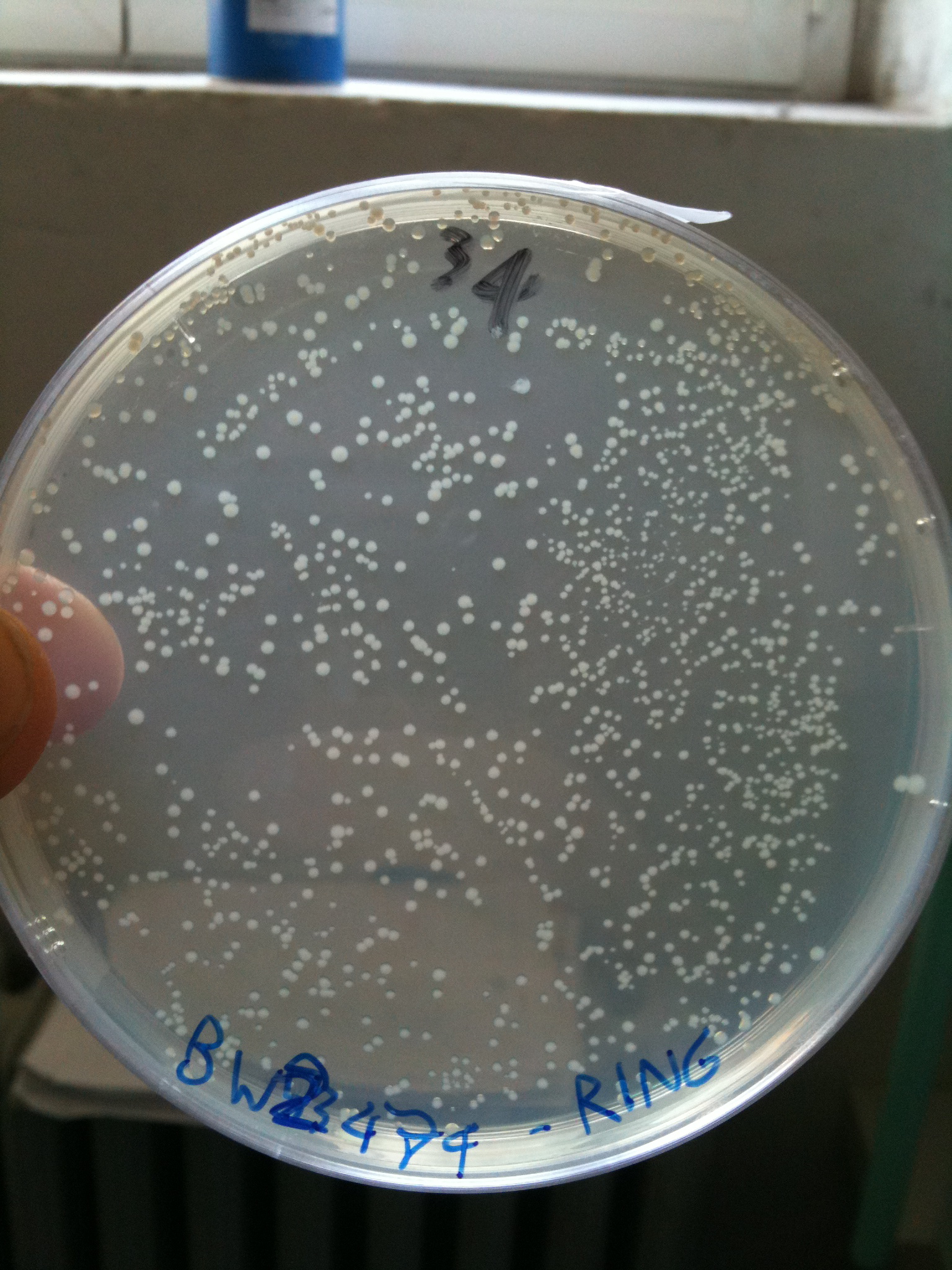 BW23474 transformed with RING |  BW25141 transformed with RING | 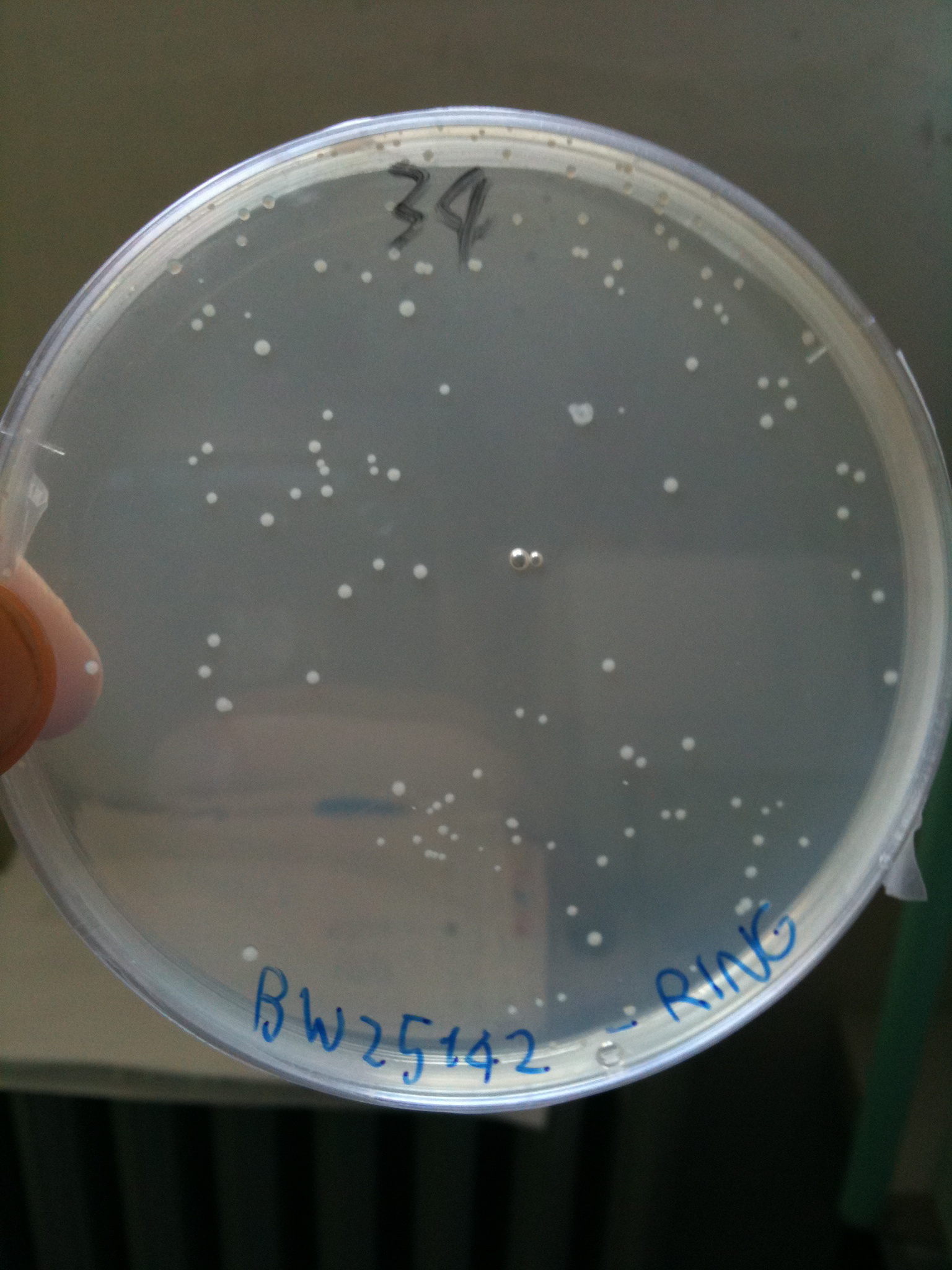 BW25142 transformed with RING |
 DH5alpha transformed with RING |  MG1655 transformed with RING |
Single colonies were picked from plates and grown in LB+Cm at proper concentration. MG1655 colonies were let grow in LB+Cm to check if they integrated the Cm resistance of RING.
MiniPrep was performed on cultures incubated yesterday, with following yields:
| Culture | Quantification
|
| I10-1 | 117.8 ng/ul
|
| I12-2 | 105,9 ng/ul
|
| I3-1 | 166,0 ng/ul
|
| I14-1 | 116,8 ng/ul
|
| I17-1 | 189,5 ng/ul
|
| <partinfo>BBa_J23110</partinfo> | 169,9 ng/ul
|
We retrieved from our freezer the following MiniPreps, with quantifications:
| Culture | Quantification
|
| 4C5 | 276 ng/ul
|
| I16-1 | 68,4 ng/ul
|
| I18-1 | 63,6 ng/ul
|
| I19-1 | 58,8 ng/ul
|
Digestion of:
| Culture | Kind | Final reaction volume (ul) | DNA (ul) | H20 (ul) | Enzyme 1 | Enzyme 2 | Buffer H
|
| <partinfo>BBa_J23110</partinfo> | Vector | 25 | 6 | 14,5 | 1 SpeI | 1 PstI | 2,5
|
| I3-1 | Vector | 25 | 10,5 | 10 | 1 XbaI | 1 PstI | 2,5
|
| 4C5 | Vector | 25 | 3,6 | 16,9 | 1 EcoRI | 1 PstI | 2,5
|
| I14-1 | Insert | 25 | 12,8 | 7,7 | 1 EcoRI | 1 PstI | 2,5
|
| I16-1 | Insert | 25 | 12,5 | 8 | 1 EcoRI | 1 PstI | 2,5
|
| I17-1 | Insert | 25 | 8 | 12,5 | 1 EcoRI | 1 PstI | 2,5
|
| I18-1 | Insert | 25 | 13 | 7,5 | 1 EcoRI | 1 PstI | 2,5
|
| I19-1 | Insert | 25 | 13 | 7,5 | 1 EcoRI | 1 PstI | 2,5
|
| I12-2 | Insert | 25 | 14 | 6,5 | 1 EcoRI | 1 PstI | 2,5
|
| I10-1 | Insert | 25 | 12,5 | 8 | 1 EcoRI | 1 PstI | 2,5
|
These parts were gel run/cut:
| I3-1 (X-P) | 5,9 ng/ul
|
| <partinfo>BBa_J23110</partinfo> (S-P) | 20,2 ng/ul
|
| I14-1 (E-P) | 13,0 ng/ul
|
| I16-1 (E-P) | 5,5 ng/ul
|
| I17-1 (E-P) | 11,9 ng/ul
|
| I18-1 (E-P) | 8,4 ng/ul
|
| I19-1 (E-P) | 4,4 ng/ul
|
4C5, I12-2 and I10-1 couldn't be extracted from gel, because bands were insignificant. For this reason we decided not to perform ligations involving I12-2 and I10-1 today. Luckily we retrieved from our freezer 4C5(E-P), so we could perform following ligations:
- I15new=<partinfo>BBa_J23110</partinfo> (S-P) + I3-1 (X-P)
- I14_4C5=I14(E-P)+4C5(E-P)
- I16_4C5=I16(E-P)+4C5(E-P)
- I17_4C5=I17(E-P)+4C5(E-P)
- I18_4C5=I18(E-P)+4C5(E-P)
- I19_4C5=I19(E-P)+4C5(E-P)
Ligations were incubated overnight at 16°C.
Glycerol stock for BW23473.
Our glycerol was contaminated, so we decided to prepare it again!
BW23474-RING was re-inoculated because it is still not grown.
We incoulated from glycerol stock (3ul in 2ml LB+Amp):
- <partinfo>BBa_J23110</partinfo>
- <partinfo>BBa_J23118</partinfo>
- <partinfo>BBa_J23116</partinfo>
- <partinfo>BBa_J23114</partinfo>
- <partinfo>BBa_J23106</partinfo>
- <partinfo>BBa_J23105</partinfo>
- <partinfo>BBa_J23101</partinfo>
- <partinfo>BBa_J23100</partinfo>
- <partinfo>BBa_B0033</partinfo>
in order to perform a TECAN test tomorrow.
Inoculum of MC1061 in LB, PBHR68 in LB+Amp and <partinfo>BBa_T9002</partinfo> in LB+Amp to prepare competent cells tomorrow.
July, 21st
Tecan Test
This morning all cultures were grown:
- BW25141-RING
- BW25142-RING
- BW23474-RING
- BW23474-RING re.inoculated yesterday night (thrown away because unuseful)
- MG1655-1
- MG1655-2
- MG1655-3
- MG1655-4
For these 7 seven cultures glycerol stocks were prepared and stored at -80°C. Remaning 5 ml were used to perform MiniPrep.
After MiniPrep, purified DNA was quantified with NanoDrop.
| Culture | Quantification
|
| BW25141-RING | 20,9 ng/ul
|
| BW25142-RING | 31,8 ng/ul
|
| BW23474-RING | 36,5 ng/ul
|
| MG1655-1 | 3,5 ng/ul
|
| MG1655-2 | 24 ng/ul
|
| MG1655-3 | 10,6 ng/ul
|
| MG1655-4 | 25,7 ng/ul
|
Digestion of:
| Culture | Kind | Final reaction volume (ul) | DNA (ul) | H20 (ul) | Enzyme 1 | Enzyme 2 | Buffer B
|
| MG1655-1 | Screening | 25 | 20 | 0,5 | 1 HindIII | 1 HindIII | 2,5
|
| MG1655-2 | Screening | 25 | 10 | 10,5 | 1 HindIII | 1 HindIII | 2,5
|
| MG1655-3 | Screening | 25 | 20 | 0,5 | 1 HindIII | 1 HindIII | 2,5
|
| MG1655-4 | Screening | 25 | 10 | 10,5 | 1 HindIII | 1 HindIII | 2,5
|
| BW25141-RING | Screening | 25 | 10 | 10,5 | 1 HindIII | 1 HindIII | 2,5
|
| BW25142-RING | Screening | 25 | 8 | 12,5 | 1 HindIII | 1 HindIII | 2,5
|
| BW23474-RING | Screening | 25 | 8 | 12,5 | 1 HindIII | 1 HindIII | 2,5
|
<partinfo>BBa_J23116</partinfo>
positive control | Screening | 25 | 2 | 18,5 | 1 HindIII | 1 HindIII | 2,5
|
Today we also prepared competent cells for cultures inoculated yesterday:
- <partinfo>BBa_T9002</partinfo> (already resistent to Ampicillin, grown in LB+Amp)
- MC1061 (grown in LB with no antibiotic)
- PBHR68 (already resistent to Ampicillin, grown in LB+Amp)
In the afternoon, we transformed our ligations:
Trasformation of ligations in:
| Ligation name | E. coli strain | Resistance |
|---|
- I15new=<partinfo>BBa_J23110</partinfo> (S-P) + I3-1 (X-P)
|
DH5alpha
|
Amp 100 |
- I14_4C5=I14(E-P)+4C5(E-P)
|
DH5alpha
|
Amp 100+Cm 12,5 |
- I16_4C5=I16(E-P)+4C5(E-P)
|
DH5alpha
|
Amp 100 + Cm 12,5 |
- I17_4C5=I17(E-P)+4C5(E-P)
|
DH5alpha
|
Amp 100 + Cm 12,5 |
- I18_4C5=I18(E-P)+4C5(E-P)
|
DH5alpha
|
Amp 100 + Cm 12,5 |
- I19_4C5=I19(E-P)+4C5(E-P)
|
DH5alpha
|
Amp 100 + Cm 12,5 |
Plates were incubated overnight at 37°C 220 rpm.
July, 22nd
plates results were not nice: I15 new showed few colonies (about 20 colonies), I14_4C5 showed 2 colonies, I16_4C5 showed 3 colonies and I18_4C5 1 colony, while for other plates no colony was observed, probably due to the fact that our 4C5 (E-P) plasmid retrieved from freezer was too old and not usable anymore. For this reason, we decided to perform a colony PCR on available colonies of I14_4C5, I16_4C5 and I18-4C5, on 3 colonies of I15new and to repeat wrong ligations and the one including I10-1 and I12-2 (their vector had to be changed with pSB4C5).
Colonies were saved in 1ml LB+suitable antibiotic/s. After gel results positive colonies will be glycerol stocked.
 Colony PCR of colonies peaked from plates Colony PCR and gel run showed that I15-1, I15-2 and I15-3 have the right insert. Also for I14_4C5-1 and I14_4C5-2 the insert was visible. For these parts we decided to perform a NheI-PstI screening. I16-4C5-1 and I16_4C5-3 showed the insert, so they will be screened as before. No band was observed for I16_4C5-2 and I18_4C5-1 (This ligation will be repeated).
Glycerol stok was prepared for I14_4C5-1, I14_4C5-2, I16_4C5-1, I16_4C5-3, I15new-1, I15new-2 and I15new-3. Falcon tubes containing the remaining culture were re-filled with 5ml LB+antibiotic and tomorrow will be screened.
Other parts (I14 E-P, I16 E-P, I17 E-P, I18E-P, I19 E-P) were available in the freezer, so only 3 parts still had to be digested.
Digestion of pSB4C5 (E-P), I12-2(E-P) and I10-1(E-P) was repeated as yesterday, digestions were gel run/cut and bands were visible, so this time gel extraction was performed with the following quantifications:
- pSB4C5 (E-P): 23,0 ng/ul
- I10-1 (E-P): 14,1 ng/ul
- I12-2 (E-P): 21,9 ng/ul
Following ligations were performed:
- I17_4C5=I17(E-P)+4C5(E-P)
- I18_4C5=I18(E-P)+4C5(E-P)
- I19_4C5=I19(E-P)+4C5(E-P)
- I10-4C5=I10(E-P)+4C5(E-P)
- I12-4C5=I12(E-P)+4C5(E-P)
In the afternoon, competent T9002 (on LB+Amp+Cm 12,5 LB agar plates) and MC1061 (on LB+Cm 12,5 agar plates) cells were transformed with a negative control (ENTERO 4C5) in pSB4C5, in order to evaluate transformation efficiency.
Plates were incubated at 37°C overnight.
We also performed a TECAN test in order to evaluate the ranking of sternght of promoters we inoculaed yesterday. Results how that the ranking is the one reported here (from stronger to weaker):
- <partinfo>BBa_J23100</partinfo>
- <partinfo>BBa_J23101</partinfo>
- <partinfo>BBa_J23110</partinfo>
- <partinfo>BBa_J23118</partinfo>
- <partinfo>BBa_J23106</partinfo>
- <partinfo>BBa_J23105</partinfo>
- <partinfo>BBa_J23116</partinfo>
- <partinfo>BBa_J23114</partinfo>
Today we performed PCR in order to amplify the phasin PhaP (<partinfo>BBa_K208001</partinfo>). Out primers are built to eliminate the stop codon from phasin and to give it the prefix and suffix we desire:
July, 23rd
July, 24th
Efficiency of transformation:
| Culture | Colonies
|
| BW25141 | 232 colonies
|
| BW25142 | 137 colonies
|
| BW23474 | 1436 colonies
|
July, 25th
|
|
 "
"








