Team:Groningen
From 2010.igem.org
Joelkuiper (Talk | contribs) |
Joelkuiper (Talk | contribs) |
||
| Line 23: | Line 23: | ||
<li> | <li> | ||
<div id="textWrapper"> | <div id="textWrapper"> | ||
| - | + | </html>{{:Team:Groningen/Images}}<html> | |
| - | </html>{{:Team:Groningen/ | + | |
</div> | </div> | ||
</li> | </li> | ||
Revision as of 13:54, 3 August 2010
- Home
- Project Overview | Biofilm | Chaplins | Expression | Applications | Biobricks
- Modeling Overview | Biofilm dynamics | Expression | Kill Switch | Information standard
- Human Practices Overview | Education | Safety | Survey | Ethics | For Parents
- Organization Overview | Brainstorm | Flow Chart | Protocols | Notebook
- Team Overview | Members | Collaboration | Image of Science | Contact
- Judging
-
Welcome to iGEM Groningen!
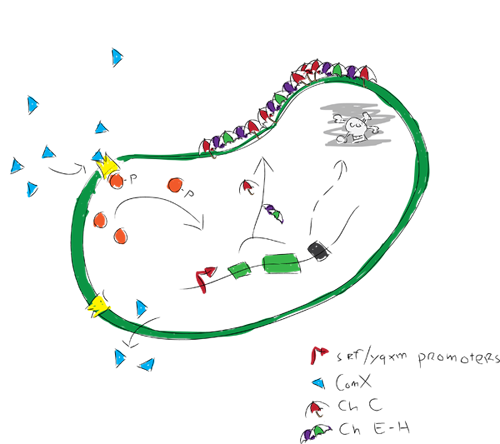
A self assembling bio-based coating is form, a rigid biofilm.Expression of hydrophobic proteins called chaplins is induced by the biofilm causing strong surface hydrophobicity.The strongly hydrophobic biofilm will die off by a killswitch, leaving a nice hydrophobic biological coating.Self assembling hydrophobic biofilm
We aim to design a biological coating as an alternative to for example chemical coatings. For this we, unconventionally, utilized a Bacillus subtilis biofilm. We wanted to enable our biofilm to be equipped with an interesting property which is automatically initiated. So we introduced an expression trigger which relies on quorum sensing. Our project was directed at finding an alternate solution to biofouling, since regular, chemical coatings which are widely in use pose a threat to the environment. In nature the lotus leaves show self-cleansing properties ascribed to their extreme surface hydrophobicity. In the prokaryotic domain we stumbled upon chaplins, strongly hydrophobic proteins originating from Streptomyces coelicolor. Surface hydrophobicity is a very useful property and is used in many applications ranging from not only antifouling coatings but also other applications which require water repellence to applications in the field of medical sciences. During our project we have contributed to the parts registry whit numerous BioBricks.
We believe that there is a great future for biological coatings as demonstrated within the project. Hydrophobic biological coatings can provide a greener antifouling solution. However using the underlying mechanisms of biofilm triggered expression for other systems like dynamic painting, sensing of environmental changes or even the integration with silicon chips might be within the realm of possibilities. So we challenge coming iGEM teams to further explore and brainstorm about the possibilities of using biofilms as a host for a wide range of applications. This year a team of young inspired undergraduates from the [http://www.rug.nl University of Groningen] participated in the amazing challenge of iGEM. A multi-disciplinary team of Molecular Biologists, Chemists, Computer Scientists, Journalists and others spend the summer creating a wonderful project in the emerging field of synthetic biology
This year a team of young inspired undergraduates from the [http://www.rug.nl University of Groningen] participated in the amazing challenge of iGEM. A multi-disciplinary team of Molecular Biologists, Chemists, Computer Scientists, Journalists and others spend the summer creating a wonderful project in the emerging field of synthetic biology Using computer models we worked on the frontiers of knowledge. Gene expression was simulated and a simple explanation for cell differentiation was proposed. Also aiding in ethics and practical feasibility a kill switch system was studied. Finally a new standard was proposed for characterizing Biobrick parts so future can be streamlined.
Using computer models we worked on the frontiers of knowledge. Gene expression was simulated and a simple explanation for cell differentiation was proposed. Also aiding in ethics and practical feasibility a kill switch system was studied. Finally a new standard was proposed for characterizing Biobrick parts so future can be streamlined.
Our sponsors
-
Self assembling hydrophobic biofilm
Self assembling hydrophobic biofilm
Surface hydrophobicity is a very useful property and is used in many applications ranging from raincoats, antifouling coatings, anti-bacterial and anti-fungal coatings and applications in the field of biomedical sciences to protection of highly sensitive sensor equipment. Hydrophobicity keeps a surface water free and thereby clean and dry, this prevents micro-organisms from fouling surfaces and corrosion from forming. Most hydrophobic coatings used today are either costly or contain harmful chemicals. So why not create an organism that does the work for you.
The idea is to engineer a bacterium that once applied on a surface, starts forming a rigid biofilm. Completion of the biofilm will trigger the expression of hydrophobic proteins by the biofilm forming bacterium. These hydrophobic proteins will be incorporated in the rigid biofilm, causing strong hydrophobic surface activity. After the hydrophobic biofilm production, a kill switch would be activated to kill of all bacteria. The result of these processes will be a surface that is coated by a rigid biofilm with embedded hydrophobic proteins, leaving the coated surface extremely hydrophobic.
Producing a hydrophobic biocoating that is self assembling, would have a lot of advantages. Firstly it is relatively cheap to apply bacteria to surfaces. And since the coating process will be done by the bacteria themselves, there is no high-tech treatment involved and there are no expensive chemicals necessary to attain the hydrophobicity. Secondly, because these hydrophobins are proteins, they are in contrast to many chemical hydrophobins non-toxic to the environment. Applications of this hydrophobic biofilm could range from antifouling coatings on ships, antifungal coatings and corrosion and water protecting coatings.
-
== Image of Science ==

Image of Science by iGEM Groningen & Joël Kuiper is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License.
Also check out the fungi we've grown during the project in our fungal gallery!
-
Modelling
Modeling
Modeling in iGEM
Together with the rise of synthetic biology, more and more computational tools and methods to improve and model biological systems were developed. Biological systems are often very complicated networks and it can be hard to study their behaviour without the help of computers. This is why mathematical descriptions of biochemical processes have become an important way of studying biological systems aside from wetwork. Computer simulations can for example predict system behaviour or screen for possible drug targets. Models can also show were the gaps in our understanding of a biological system are, because they might show that the assumed mechanism is not right.
In synthetic biology, modelling is important because it can save time and money and improve the final product. Mathematical models do have to be validated by comparing experimental and simulation results, this is why models have become a tool for use in parallel with experimental approaches. However, modeling in synthetic biology is an interdisciplinary task, because one needs to understand both the biology of the systems which will be modeled and posess the mathematical knowledge needed to build the model. Therefore, modelling is a collaborative effort of different fields of science.
Our modeling goals
When it became clear that our project involved biofilms, we looked into models that describe biofilm formation. We found that such models existed and visited an expert on biofilm models in Delft. After this visit, it became clear that although these models are pretty mathematical, we could make a simple model with biofilm formation and diffusion.
We modeled gene expression of our hydrofobic proteins. The modelers were actually the ones who discovered the qurom sensing pathway used in our project when looking in the partsregistry. The Com XPA system has since then been studied and we found that the biological pathways involved are far more complex than our model could describe. We therefore chose to model how the ComXPA system could activate the expression of our proteins.
We also looked into killswitch mechanisms because the applications of our biofilm in for example anti-fouling coatings are outside the lab. Since most GMO's are not allowed to leave the lab alive, it would be nice to kill our bacteria after they formed a biofilm with our hydrofobic proteins. Therefore we modeled a simple toxin anti-toxin killswitch mechanism.
Finally we wrote an information standard which hopefully provides more and easier searchable information for the partsregistry.
References
Lukas Endler, Nicolas Rodriguez, Nick Juty, Vijayalakshmi Chelliah, Camille Laibe, Chen Li and Nicolas Le Novère, Designing and encoding models for synthetic biology, Journal of the Royal Society Interface, 2009, online April 1, http://rsif.royalsocietypublishing.org/content/early/2009/03/27/rsif.2009.0035.focus.full. Yiannis N Kaznessis, Models for synthetic biology, BMC Systems Biology 2007, 1:47, http://www.biomedcentral.com/1752-0509/1/47.
-
Our Team
== Team ==(f.l.t.r: Ramon, Arend Jan, Peter, Jorrit, Geeske, Maarten, David, Arend, Djoke, Joël)
Go check out the members of our team, with whom we collaborated or our publicly available pictures! If you still have questions you can always contact us!
Our advisors
- prof. dr. Oscar Kuipers: [http://molgen.biol.rug.nl/molgen/index.php Molecular Genetics] (Head)
- dr. Dennis Claessen: [http://www.rug.nl/staff/d.claessen/index Microbial Physiology]
- prof. dr. Jan Kok: [http://molgen.biol.rug.nl/molgen/index.php Molecular Genetics]
- prof. dr. Bert Poolman: Biochemistry; [http://www.centreforsyntheticbiology.eu/ Centre for Synthetic Biology] (Director)
- prof. dr. Roel Bovenberg: Synthetic biology and Cell engineering; Corporate Scientist Biotechnology, [http://www.dsm.com/ DSM]
-
Contact us
Contact us
University of Groningen iGEM Team
Biological Center
Email: igem@igemgroningen.com
Website: [http://www.igemgroningen.com igemgroningen.com]
Telephone: +31503632276
Kerklaan 30
Postbus 14
9750 AA Haren
The Netherlands
Feel free to contact us!
 "
"