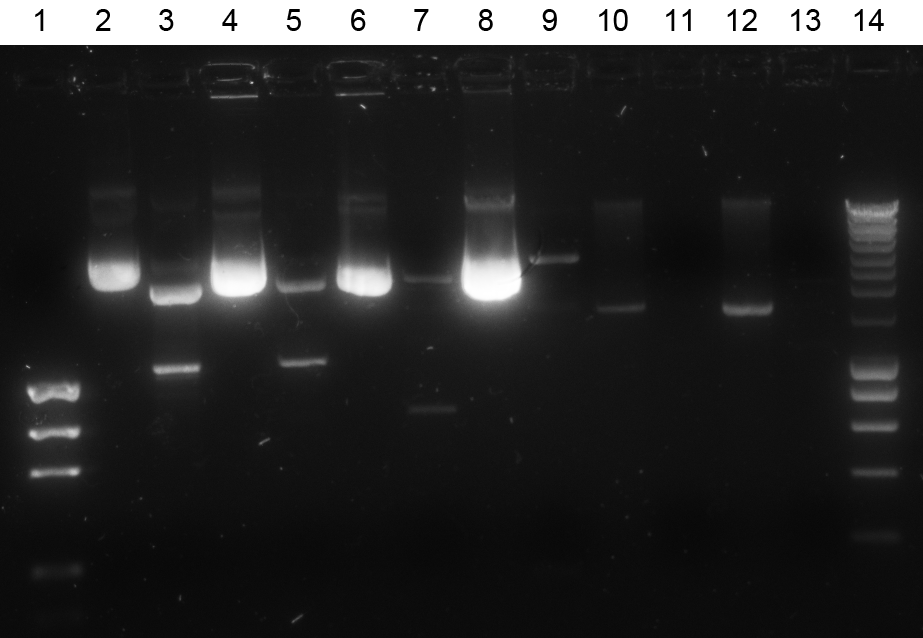Team:TU Delft/13 July 2010 content
From 2010.igem.org
(Difference between revisions)
(→Characterization of Anderson RBS sequences) |
(→Lab work) |
||
| Line 192: | Line 192: | ||
'''!''' Sample not fully loaded on gel | '''!''' Sample not fully loaded on gel | ||
| + | |||
| + | The band position of the digestion products of AlnA and OprG correspond to their length. The other fragments (R0011, B0032 and B0015) probably run of the gel. | ||
| + | |||
| + | The ligation products that were incubated over night were transformed to Top10 competent cells according to the [[Team:TU_Delft/protocols/transformation|protocol]]. | ||
| + | |||
==Characterization of Anderson RBS sequences== | ==Characterization of Anderson RBS sequences== | ||
Revision as of 19:51, 19 July 2010
Contents |
Lab work
Ordered DNA
We have now stock of the ordered DNA, to make a real BioBrick of this DNA we are going to ligated it into the iGEM plasmid backbone SB1C3. First we digested the plasmids:
| # | Digestion reaction | Used Buffer | Needed fragment |
| 1 | 1 μg alkB2+ EcoRI + PstI | Buffer 3 (BioLabs) | ‘E – alkB2– P’ |
| 2 | 1 μg rubA3+ SpeI + PstI | Buffer 3 (BioLabs) | ‘E – rubA3– P’ |
| 3 | 1 μg ladA + SpeI + PstI | Buffer 3 (BioLabs) | ‘E – ladA – P’ |
| 4 | 1 μg ADH + SpeI + PstI | Buffer 3 (BioLabs) | ‘E – ADH – P’ |
| 5 | 1 μg AlnA + SpeI + PstI | Buffer 3 (BioLabs) | ‘E – AlnA – P’ |
| 6 | 1 μg OprG + SpeI + PstI | Buffer 3 (BioLabs) | ‘E – OprG – P’ |
| 7 | 1 μg AlkS + SpeI + PstI | Buffer 3 (BioLabs) | ‘E – AlkS – P’ |
| 8 | 1 μg PalkB + SpeI + PstI | Buffer 3 (BioLabs) | ‘E – PalkB – P’ |
| 9 | 1 μg PalkS12+ SpeI + PstI | Buffer 3 (BioLabs) | ‘E – PalkS12– P’ |
| 10 | 1 μg PhPFDα + SpeI + PstI | Buffer 3 (BioLabs) | ‘E – PhPFDα – P’ |
| 11 | 1 μg PhPFDβ + SpeI + PstI | Buffer 3 (BioLabs) | ‘E – PhPFDβ – P’ |
| 12 | 3 μg pSB1C3 + EcoRI + PstI | Buffer 3 (BioLabs) | ‘E – ---- – P’ |
Solvent Tolerance and Hydrocarbon Sensing
Some BioBricks are in production. Digestion reaction
| # | Digestion reaction | Used Buffer | Needed fragment |
| 1 | 1 μg PhPFDα + EcoRI + SpeI | Buffer 2 + BSA (BioLabs) | ‘E – PhPFDα – S’ |
| 2 | 1 μg PhPFDβ + EcoRI + SpeI | Buffer 2 + BSA (BioLabs) | ‘E – PhPFDβ – S’ |
| 3 | 1 μg AlkS + EcoRI + SpeI | Buffer 2 + BSA (BioLabs) | ‘E – AlkS – S’ |
| 4 | 1 μg PalkB + EcoRI + SpeI | Buffer 2 + BSA (BioLabs) | ‘E – PalkB – S’ |
| 5 | 1 μg B0032 + XbaI + PstI | Buffer 2 + BSA (BioLabs) | ‘X – B0032 – P’ |
| 6 | 1 μg B0015 + XbaI + PstI | Buffer 2 + BSA (BioLabs) | ‘X – B0015 – P’ |
| 7 | 1 μg E0422 + XbaI + PstI | Buffer 2 + BSA (BioLabs) | ‘X – E0422 – P’ |
| 8 | 3 μg pSB1T3 + EcoRI + PstI | Buffer 2 + BSA (BioLabs) | ‘E - ---- - P’ |
Emulsifier
Today the digestion products from yesterday were run on gel to see whether the plasmids were cut in the right way.
Lane description
| # | Description | Expected Length (bp) |
| 1 | Biorad EZ marker (5 μL) | |
| 2 | Undigested pSB1T3 (10 μL + 2 μL loadingbuffer) | |
| 3 | Digested pSB1T3 (10 μL + 2 μL loadingbuffer) | 2500 |
| 4 | Undigested AlnA (10 μL + 2 μL loadingbuffer) | |
| 5 | Digested AlnA (10 μL + 2 μL loadingbuffer) | 1107 |
| 6 | Undigested OprG (10 μL + 2 μL loadingbuffer) | |
| 7 | Digested OprG (10 μL + 2 μL loadingbuffer) | 744 |
| 8 | Undigested B0015 (10 μL + 2 μL loadingbuffer) | |
| 9 | Digested B0015 (10 μL + 2 μL loadingbuffer) | 130 |
| 10 | Undigested R0011 (10 μL + 2 μL loadingbuffer) ! | |
| 11 | Digested R0011 (10 μL + 2 μL loadingbuffer) | 55 |
| 12 | Undigested B0032 (10 μL + 2 μL loadingbuffer) | |
| 13 | Digested B0032 (10 μL + 2 μL loadingbuffer) | 13 |
| 14 | SmartLadder (5 μL) |
! Sample not fully loaded on gel
The band position of the digestion products of AlnA and OprG correspond to their length. The other fragments (R0011, B0032 and B0015) probably run of the gel.
The ligation products that were incubated over night were transformed to Top10 competent cells according to the protocol.
 "
"