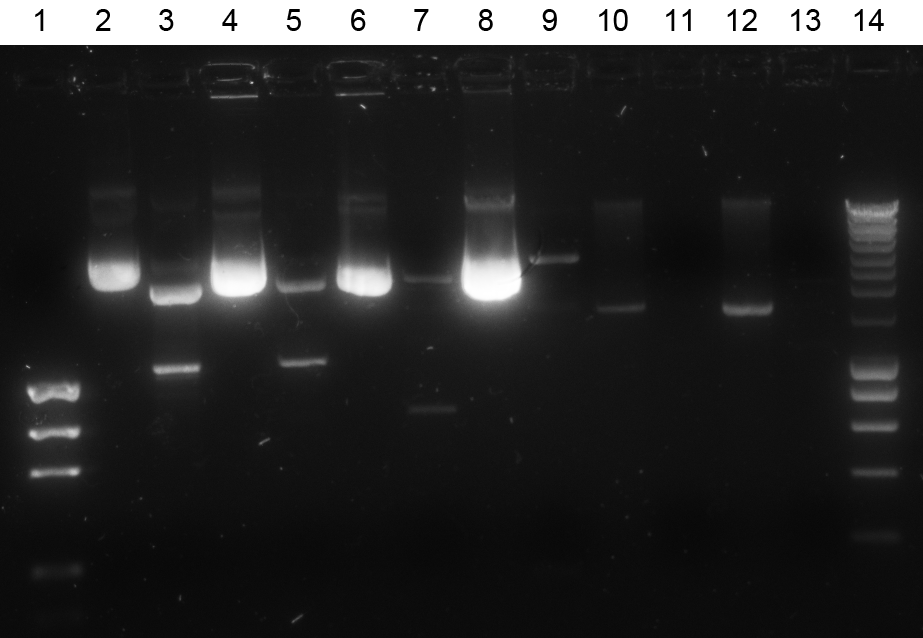Team:TU Delft/13 July 2010 content
From 2010.igem.org
(Difference between revisions)
(→Lab work) |
(→Characterization of Anderson RBS sequences) |
||
| Line 193: | Line 193: | ||
'''!''' Sample not fully loaded on gel | '''!''' Sample not fully loaded on gel | ||
==Characterization of Anderson RBS sequences== | ==Characterization of Anderson RBS sequences== | ||
| - | |||
| - | |||
| - | |||
Revision as of 19:50, 19 July 2010
Contents |
Lab work
Ordered DNA
We have now stock of the ordered DNA, to make a real BioBrick of this DNA we are going to ligated it into the iGEM plasmid backbone SB1C3. First we digested the plasmids:
| # | Digestion reaction | Used Buffer | Needed fragment |
| 1 | 1 μg alkB2+ EcoRI + PstI | Buffer 3 (BioLabs) | ‘E – alkB2– P’ |
| 2 | 1 μg rubA3+ SpeI + PstI | Buffer 3 (BioLabs) | ‘E – rubA3– P’ |
| 3 | 1 μg ladA + SpeI + PstI | Buffer 3 (BioLabs) | ‘E – ladA – P’ |
| 4 | 1 μg ADH + SpeI + PstI | Buffer 3 (BioLabs) | ‘E – ADH – P’ |
| 5 | 1 μg AlnA + SpeI + PstI | Buffer 3 (BioLabs) | ‘E – AlnA – P’ |
| 6 | 1 μg OprG + SpeI + PstI | Buffer 3 (BioLabs) | ‘E – OprG – P’ |
| 7 | 1 μg AlkS + SpeI + PstI | Buffer 3 (BioLabs) | ‘E – AlkS – P’ |
| 8 | 1 μg PalkB + SpeI + PstI | Buffer 3 (BioLabs) | ‘E – PalkB – P’ |
| 9 | 1 μg PalkS12+ SpeI + PstI | Buffer 3 (BioLabs) | ‘E – PalkS12– P’ |
| 10 | 1 μg PhPFDα + SpeI + PstI | Buffer 3 (BioLabs) | ‘E – PhPFDα – P’ |
| 11 | 1 μg PhPFDβ + SpeI + PstI | Buffer 3 (BioLabs) | ‘E – PhPFDβ – P’ |
| 12 | 3 μg pSB1C3 + EcoRI + PstI | Buffer 3 (BioLabs) | ‘E – ---- – P’ |
Solvent Tolerance and Hydrocarbon Sensing
Some BioBricks are in production. Digestion reaction
| # | Digestion reaction | Used Buffer | Needed fragment |
| 1 | 1 μg PhPFDα + EcoRI + SpeI | Buffer 2 + BSA (BioLabs) | ‘E – PhPFDα – S’ |
| 2 | 1 μg PhPFDβ + EcoRI + SpeI | Buffer 2 + BSA (BioLabs) | ‘E – PhPFDβ – S’ |
| 3 | 1 μg AlkS + EcoRI + SpeI | Buffer 2 + BSA (BioLabs) | ‘E – AlkS – S’ |
| 4 | 1 μg PalkB + EcoRI + SpeI | Buffer 2 + BSA (BioLabs) | ‘E – PalkB – S’ |
| 5 | 1 μg B0032 + XbaI + PstI | Buffer 2 + BSA (BioLabs) | ‘X – B0032 – P’ |
| 6 | 1 μg B0015 + XbaI + PstI | Buffer 2 + BSA (BioLabs) | ‘X – B0015 – P’ |
| 7 | 1 μg E0422 + XbaI + PstI | Buffer 2 + BSA (BioLabs) | ‘X – E0422 – P’ |
| 8 | 3 μg pSB1T3 + EcoRI + PstI | Buffer 2 + BSA (BioLabs) | ‘E - ---- - P’ |
Emulsifier
Today the digestion products from yesterday were run on gel to see whether the plasmids were cut in the right way.
Lane description
| # | Description | Expected Length (bp) |
| 1 | Biorad EZ marker (5 μL) | |
| 2 | Undigested pSB1T3 (10 μL + 2 μL loadingbuffer) | |
| 3 | Digested pSB1T3 (10 μL + 2 μL loadingbuffer) | 2500 |
| 4 | Undigested AlnA (10 μL + 2 μL loadingbuffer) | |
| 5 | Digested AlnA (10 μL + 2 μL loadingbuffer) | 1107 |
| 6 | Undigested OprG (10 μL + 2 μL loadingbuffer) | |
| 7 | Digested OprG (10 μL + 2 μL loadingbuffer) | 744 |
| 8 | Undigested B0015 (10 μL + 2 μL loadingbuffer) | |
| 9 | Digested B0015 (10 μL + 2 μL loadingbuffer) | 130 |
| 10 | Undigested R0011 (10 μL + 2 μL loadingbuffer) ! | |
| 11 | Digested R0011 (10 μL + 2 μL loadingbuffer) | 55 |
| 12 | Undigested B0032 (10 μL + 2 μL loadingbuffer) | |
| 13 | Digested B0032 (10 μL + 2 μL loadingbuffer) | 13 |
| 14 | SmartLadder (5 μL) |
! Sample not fully loaded on gel
 "
"