Team:Lethbridge/Notebook/Lab Work/July
From 2010.igem.org
(→July 5/2010) |
JustinVigar (Talk | contribs) (→July 5/2010) |
||
| Line 65: | Line 65: | ||
<tr><td>5</td><td>4 - SRBS (A7)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | <tr><td>5</td><td>4 - SRBS (A7)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | ||
<tr><td>6</td><td>5 - CFP Complete (A8)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | <tr><td>6</td><td>5 - CFP Complete (A8)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | ||
| - | <tr><td>7</td><td>6 - SRBS</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | + | <tr><td>7</td><td>6 - SRBS (A10)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> |
| - | <tr><td>8</td><td>7 - EYFP</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | + | <tr><td>8</td><td>7 - EYFP (B1)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> |
| - | <tr><td>9</td><td></td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | + | <tr><td>9</td><td>8 - N term tag (B2)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> |
| - | <tr><td>10</td><td></td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | + | <tr><td>10</td><td>9 - pSB NEYFP (B4)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> |
| - | <tr><td>11</td><td></td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | + | <tr><td>11</td><td>10 - pSB NEYFP (B5)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> |
| - | <tr><td>12</td><td></td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | + | <tr><td>12</td><td>11 - CFP (B6)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> |
| - | <tr><td>13</td><td></td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | + | <tr><td>13</td><td>12 - pBAD-TetR (B10)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> |
| - | <tr><td>14</td><td></td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | + | <tr><td>14</td><td>13 - D3</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> |
| - | <tr><td>15</td><td></td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | + | <tr><td>15</td><td>14 - C term (D4)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> |
| - | <tr><td>16</td><td></td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | + | <tr><td>16</td><td>15 - C term (D5)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> |
| - | <tr><td>17</td><td></td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | + | <tr><td>17</td><td>16 - pLacI (D6)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> |
| - | + | <tr><td>18</td><td>17 - NEYFP (E2)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | |
| + | <tr><td>19</td><td>18 - CEYFP (E6)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>20</td><td>19 - CEYFP (E7)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>1</td><td>1kb ladder</td><td>0.5 Ladder + 2 Dye (6X) + 7.5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>2</td><td>20 - EYFP (E8)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>3</td><td>21 - EYFP (E9)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>4</td><td>22 - EYFP (E10)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>5</td><td>23 - ECFP (F1)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>6</td><td>24 - ECFP (F2)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>7</td><td>25 - ECFP (F3)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>8</td><td>26 - pBAD-TetR (F4)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>9</td><td>27 - pBAD-TetR (F5)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>10</td><td>28 - EYFP (G1)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>11</td><td>29 - pSB CEYFP (G4)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>12</td><td>30 - pBAD (1) (G6)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>13</td><td>31 - pBAD (2) (G7)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>14</td><td>32 - N term tag (G8)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>15</td><td>33 - lumazine (G9)</td><td>1 DNA + 2 Dye (6X) + 7 Milli-Q H<sub>2</sub>O</td></tr> | ||
</table><br> | </table><br> | ||
Revision as of 22:29, 19 July 2010
Back to Notebook
Back to Lab Work
Contents |
July 2010
July 5/2010
(In Lab: JV, AV, HB)
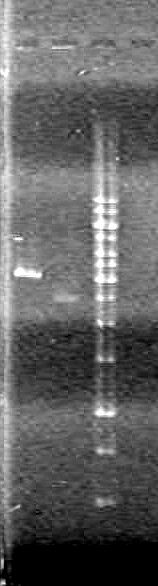
Objective: Run a 1% agarose gel of purified PCR samples from June 24/10
Method:
| Lane | Sample | Components (µL) |
| 1 | 1kb Ladder | 0.5 Ladder + 2 Dye (6X) + 7.5 Milli-Q H2O |
| 2 | 1 - pBAD (A4) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 3 | 2 - pBAD (A5) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 4 | 3 - SRBS (A6) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 5 | 4 - SRBS (A7) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 6 | 5 - CFP Complete (A8) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 7 | 6 - SRBS (A10) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 8 | 7 - EYFP (B1) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 9 | 8 - N term tag (B2) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 10 | 9 - pSB NEYFP (B4) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 11 | 10 - pSB NEYFP (B5) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 12 | 11 - CFP (B6) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 13 | 12 - pBAD-TetR (B10) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 14 | 13 - D3 | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 15 | 14 - C term (D4) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 16 | 15 - C term (D5) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 17 | 16 - pLacI (D6) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 18 | 17 - NEYFP (E2) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 19 | 18 - CEYFP (E6) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 20 | 19 - CEYFP (E7) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 1 | 1kb ladder | 0.5 Ladder + 2 Dye (6X) + 7.5 Milli-Q H2O |
| 2 | 20 - EYFP (E8) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 3 | 21 - EYFP (E9) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 4 | 22 - EYFP (E10) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 5 | 23 - ECFP (F1) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 6 | 24 - ECFP (F2) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 7 | 25 - ECFP (F3) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 8 | 26 - pBAD-TetR (F4) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 9 | 27 - pBAD-TetR (F5) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 10 | 28 - EYFP (G1) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 11 | 29 - pSB CEYFP (G4) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 12 | 30 - pBAD (1) (G6) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 13 | 31 - pBAD (2) (G7) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 14 | 32 - N term tag (G8) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
| 15 | 33 - lumazine (G9) | 1 DNA + 2 Dye (6X) + 7 Milli-Q H2O |
June 2/2010
(In Lab: JV)
Objective: Isolate plasmid DNA of RBS-xylE (BBa_J33204) from DH5α cells and confirm results.
Method: "Mini-prep" the plasmid DNA using boiling lysis miniprep. Then restrict the DNA once and run on a 1% agarose gel (TAE).
Restriction Reaction
| Ingredient | Volume(µL) |
| MilliQ H20 Water | 15.75 |
| Orange Buffer (10x) | 2 |
| pDNA (rbs-xylE) | 2 |
| EcoRI | 0.25 |
Unrestricted Control
| Ingredient | Volume(µL) |
| MilliQ H20 Water | 16 |
| Orange Buffer (10x) | 2 |
| pDNA (rbs-xylE) | 2 |
DNA was restricted for 80 minutes at 37oC.
Analyzed results on a 1% agarose gel. Load order as follows:
| Lane | Sample | Volume Sample (µL) | Volume Loading Dye (µL) |
| 1 | Restricted RBS-xylE | 10 | 2 |
| 1 | Unestricted RBS-xylE† | 1 | 2 |
| 1 | 1kb Ladder†† | 2 | 2 |
† Added 9µL MilliQ H2O
†† Added 8µL MilliQ H2O
Ran gel at 100V from 2 hours.
Results:
Conclusions: Plasmid DNA prep and restriction was successful.
Objective: Ligate rbs-xylE (Bba_J33204) to our double terminator, and insert it into the pSB1T3 plasmid backbone.
Method:
- Restrictions
- Restrict rbs-xylE wit EcoRI and SpeI (Red Buffer)
- Restrict the double terminator with XbaI and PstI (Tango Buffer)
- Restrict pSB1T3 with EcoRI and PstI (Red Buffer)
Component Volume (µL) MilliQ H2O 15.5 Buffer 2 pDNA 2 Enzyme 0.25 + 0.25 Set up control reaction as follows:
- MilliQ H2O - 16µL
- Buffer - 2µL
- pDNA - 2µL
Incubated reactions for 65 minutes at 37oC
Killed enzymes by incubating reactions for 10 minutes at 65oC
- Ligation
Reaction set up as follows:- T4 DNA ligase - 0.25µL
- rbs-xylE - 5µL
- dT - 3µL
- pSB1T3 - 8µL
- 10x Ligation Buffer - 2µL
- MilliQ H2O - 1.75µL
Killed enzymes by incubating reactions for 10 minutes at 80oC</ul>June 2/2010 - Evening
Objective: Set up new ligations of pLacI and sRBS-Lum-dT according to Tom Knight's protocol. Previous ligation had very little DNA.
Relevant Information:
- Want a final mass of 25ng of each pDNA in the ligation mix.
- Final concentration of pDNA in restriction digest should be 25-50ng/µL.
- Tom Knight's restriction reaction is 50µL, therefore there should be 1000ng pDNA in each restriction digest.
- Identified the following plasmids in our working plasmids box:
Common Name Location Concentration (ng/µL) Volume/rxn (µL) pLacI Maxiprep A9 990 ~1 pLacI (B1) A6 440 ~2 sRBS-Lum-dT (2) A1 965 ~1 sRBS-Lum-dT (1) A2 1145 ~1 sRBS-Lum-dT Maxiprep B8 4780 ~.2 sRBS-Lum-dT B7 4375 ~.25 sRBS-Lum-dT (1) G2 335 ~3 sRBS-Lum-dT (2) G3 965 ~2 - Make a 1:10 dilution of sRBS-Lum-dt maxiprep (D8) and sRBS-Lum-dT (B7). 0.5µL pDNA in 4.5µL water.
- Cut pLacI with EcoRI and SpeI
- Cut sRBS-Lum-dT with XbaI and PstI
- Cut pSB1T3 with EcoRI and PstI
- Will have total of 12 ligation reactions, want 12x2µL of pSB1T3 to add to each, therefore want 25µL of pSB1T3.
Method:
Restriction
Name [pDNA] (ng/µL) Volume
pDNA (µL)Volume
Water (µL)Volume
Buffer (µL)Enzymes Total Volume sRBS-Lum-dT (A1) 965 1 43.5 5 0.25µL XbaI
0.25µL PstI50 sRBS-Lum-dT (A2) 1145 1 43.5 5 0.25µL XbaI
0.25µL PstI50 pLacI Maxiprep (A1) 990 1 43.5 5 0.25µL EcoRI
0.25µL SpeI50 sRBS-Lum-dT Maxiprep(B8) 4780 2 (of 1:10 dilution) 42.5 5 0.25µL XbaI
0.25µL PstI50 sRBS-Lum-dT (B7) 4375 2.5 (of 1:10 dilution) 42 5 0.25µL XbaI
0.25µL PstI50 pLacI (D6) 440 2 42.5 5 0.25µL EcoRI
0.25µL SpeI50 sRBS-Lum-dT (G2) 335 3 41.5 5 0.25µL XbaI
0.25µL PstI50 sRBS-Lum-dT (G3) 540 2 42.5 5 0.25µL XbaI
0.25µL PstI50 pSB1T3 25 12.5 7 5 0.25µL EcoRI
0.25µL PstI50 Incubate for 30 minutes at 37oC (Start- 12:10pm; End- 12:40pm)
Heat kill enzymes at 80oC for 20 minutes
Ligation:
In a 10µL final volume, add:- 2µL of sRBS-Lum-dT component
- 2µL of pLacI component
- 2µL of pSB1T3 component
- 1µL of T4 Buffer
- 0.25µL of T4 DNA Ligase
- 2.75µL of MilliQ H2O
Incubate for 30 minutes at room temperature to ligate
Incubate for 20 minutes at 80oC to heat kill
- Restrict rbs-xylE wit EcoRI and SpeI (Red Buffer)
 "
"


 ]
]
 ]
]
 ]
]
 ]
]
 ]
]
 ]
]
 ]
]
 ]
]