Team:Slovenia/PROJECT/proof
From 2010.igem.org
| Line 19: | Line 19: | ||
<br> | <br> | ||
The main idea of our project was to use DNA-binding domains, e.g. zinc fingers specific for unique target elements within a predesigned DNA program sequence, and show their ability to act as anchors for various protein functional domains (e.g. enzymes and split fluorescent proteins). Our main goal was to first characterize binding properties of DNA binding factors and demonstrate that their binding could be defined by the order of DNA target elements on DNA program sequence and later use them in an application, such as biosynthesis. | The main idea of our project was to use DNA-binding domains, e.g. zinc fingers specific for unique target elements within a predesigned DNA program sequence, and show their ability to act as anchors for various protein functional domains (e.g. enzymes and split fluorescent proteins). Our main goal was to first characterize binding properties of DNA binding factors and demonstrate that their binding could be defined by the order of DNA target elements on DNA program sequence and later use them in an application, such as biosynthesis. | ||
| + | <br> | ||
In order to prove the concept described above we had to: | In order to prove the concept described above we had to: | ||
Revision as of 20:01, 27 October 2010
The main idea of our project was to use DNA-binding domains, e.g. zinc fingers specific for unique target elements within a predesigned DNA program sequence, and show their ability to act as anchors for various protein functional domains (e.g. enzymes and split fluorescent proteins). Our main goal was to first characterize binding properties of DNA binding factors and demonstrate that their binding could be defined by the order of DNA target elements on DNA program sequence and later use them in an application, such as biosynthesis.
In order to prove the concept described above we had to:
- select the appropriate DNA-binding domains,
- demonstrate their specific binding to DNA motifs,
- show that the addition of functional domain does not interfere with their binding to DNA,
- show that we can bind several adjacent DNA-binding proteins linked to functional protein domains to the DNA program and demonstrate the function of such an assembled protein
There are many proteins domains that bind to defined DNA sequences, such as restriction endonucleases, transcription factors, zinc fingers, transcriptional activator like (TAL) elements, etc.
We found zinc finger domains most appropriate for our purpose, since they can be designed to bind selected DNA sequence through concatenation of small domains called fingers that recognize DNA trimers. There are zinc fingers that recognize from 9 up to 18 nucleotides. Recently the molecular code of DNA recognition by TAL effectors was deciphered (Boch et al., 2009), which provides another tool to design sequence specific DNA recognition proteins.
Two zinc fingers were already present in the Registry: Gli1 and ZNF HIVC, however their functionality has not been shown. In order to test our idea additional zinc fingers needed to be obtained (see list and characterization of parts - [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2010&group=Slovenia link]).
We selected zinc finger proteins Tyr456, Zif268, PBSII, Blues and Jazz and order synthetic genes from GeneArt company. The first aim of our project was to show binding of zinc finger proteins to a specific DNA target sequence.
We tested binding of zinc fingers with added protein domains to DNA using several techniques: electrophoretic mobility shift analysis (EMSA), surface plasmon resonance (SPR), our new universal detection device composed of following two parts: [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323088 BBa_323088] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323089 BBa_323089] for qualitative and quantitative analysis of DNA binding properties of selected DNA binding domains in vivo based on beta-galactosidase reporter.
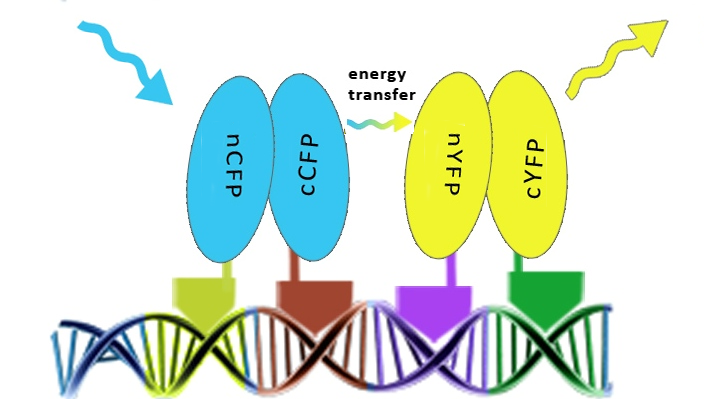
The first step providing proof of the principle was to test whether zinc fingers assemble on a DNA scaffold with the cognate binding sites using split fluorescent protein reconstitution. Fluorescent proteins were split into two overlapping segments and fused to different zinc fingers that should bind to adjacent sites on DNA program.
Once the reconstitution of pairs of fluorescent proteins was confirmed we could test binding of four zinc fingers next to each other, which led to the Förster resonance energy transfer (FRET). These experiments were performed in mammalian cells, since emmitting fluorescence in mammalian cells is relatively easy to detect with a confocal microscope.
Results on the following pages show that we characterized binding properties of 6 zinc fingers to their respective DNA elements, including two (Gli1 and HIVC) that have previously been deposited in the Registry, but not characterized. We demonstrated proof of the principle of DNA-guided assembly of four different zinc finger fusion proteins by FRET. FRET device consists of five elements: four fusion proteins with different DNA binding domains and a separate DNA program sequence. It has a potential to be used for the purpose of information processing and pattern recognition, however, other fused functional protein domains may be more versatile in comparison to the fluorescent proteins and FRET.
 "
"