Team:EPF Lausanne/Project immuno
From 2010.igem.org
(→Proteins) |
|||
| Line 43: | Line 43: | ||
Now if Asaia starts to produce a hight amout of these proteins, the interactions will be disrupted and the plasmodium will not propagate through the mosquito's epithelium. | Now if Asaia starts to produce a hight amout of these proteins, the interactions will be disrupted and the plasmodium will not propagate through the mosquito's epithelium. | ||
| + | |||
| + | |||
=Results= | =Results= | ||
==A: The Immunotoxin is expressed and appears in the supernatant== | ==A: The Immunotoxin is expressed and appears in the supernatant== | ||
| - | [[Image:EPFL Immuno growth.jpg|300px|thumb|right| <b>Growth curve of E. coli expressing the Immunotoxin:</b> In blue, we can see the growth curve of E.coli containing the empty C3 plasmid. In red, we can see the growth of E.coli transformed with the C3 plasmid containing the Immunotoxin sequence. We could conclude that the expression of the Immunotoxin in high concetrations may prevent E.coli from growing | + | [[Image:EPFL Immuno growth.jpg|300px|thumb|right| <b>Growth curve of ''E. coli'' expressing the Immunotoxin:</b> In blue, we can see the growth curve of ''E. coli'' containing the empty C3 plasmid. In red, we can see the growth of ''E. coli'' transformed with the C3 plasmid containing the Immunotoxin sequence. We could conclude that the expression of the Immunotoxin in high concetrations may prevent ''E. coli'' from growing. ]] |
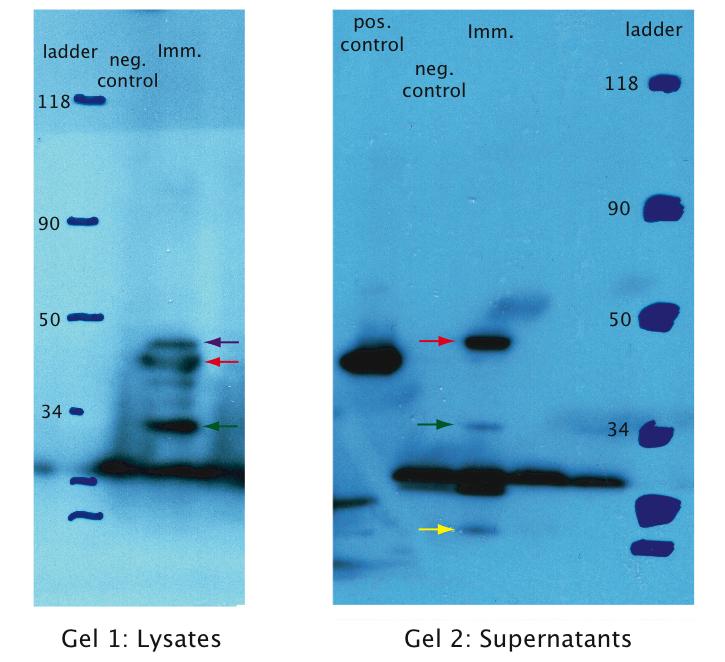
| - | We tested expression of the immunotoxin in E. | + | We tested expression of the immunotoxin in ''E. coli'' (see [https://2010.igem.org/wiki/index.php?title=Team:EPF_Lausanne/Project/Materials_Methods Materials and Methods] for details). In a western blot analysis of whole cell lysates we could see bands corresponding to full length immunotoxin and possibly degraded fragments of the protein (see figure). The immunotoxin contains a PelB sequence that targets it for secretion into the periplasm. We concentrated the supernatant of both the immunotoxin and a control culture by running it through a filtering device with a 5 kDa cut-off. Running a western blot with these samples (see figure) we verified that the immunotoxin was also found in the supernatant. |
| - | [[Image:EPFL Gel1 western.jpg|400px|thumb|center| <b>Gel1 (Lysates):</b> We can see | + | [[Image:EPFL Gel1 western.jpg|400px|thumb|center| <b>Gel1 (Lysates):</b> We can see distinct molecular fragments in the immunotoxin lane: The upper fragments (marked with red and purple arrows) match the molecular size of the immunotoxin of about 39 kDa. For the shorter of the two fragments the export tag pelB may have been cut off. The smaller sized fragment (green arrow) of about 20 kDa is most probably part of the degraded immunotoxin. |
<b>Gel2 (Supernatants):</b> A fragment of the correct size is detected in the supernatant. The smaller fragments (green and yellow arrows) probably correspond to smaller parts of the degraded immunotoxin. ]] | <b>Gel2 (Supernatants):</b> A fragment of the correct size is detected in the supernatant. The smaller fragments (green and yellow arrows) probably correspond to smaller parts of the degraded immunotoxin. ]] | ||
==B: The P-proteins are not expressed == | ==B: The P-proteins are not expressed == | ||
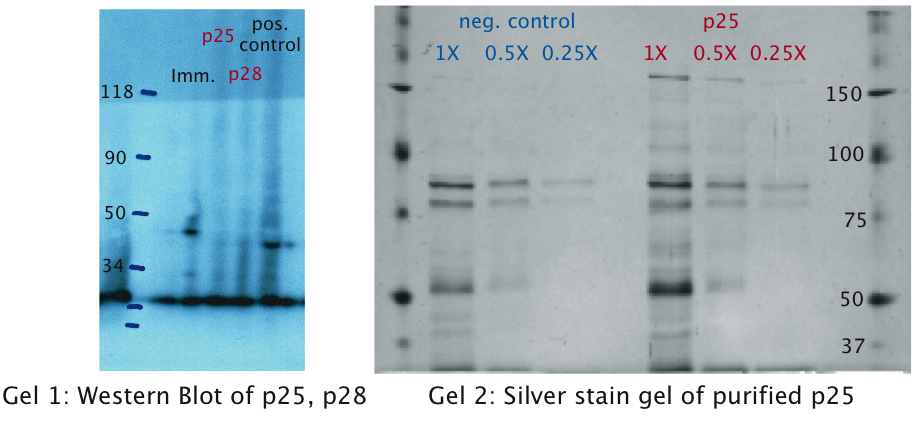
| - | The same experiments were conducted for the proteins [https://2010.igem.org/wiki/index.php?title=Team:EPF_Lausanne/Project/Background p25 and p28]. No bands were detected on the western blots (see figure) which leads us to the conclusion that these proteins were only very weakly expressed or not at all. This might be explained by the fact that we took the native sequence from <i>Plasmodium falciparum </i>. The genome of <i>Plasmodium falciparum </i> is very A-T-rich ([http://areslab.ucsc.edu/cgi-bin/hgGateway UCSC Malaria Genome Browser]). | + | The same experiments were conducted for the p-proteins [https://2010.igem.org/wiki/index.php?title=Team:EPF_Lausanne/Project/Background p25 and p28]. No bands were detected on the western blots (see figure) which leads us to the conclusion that these proteins were only very weakly expressed or not at all. This might be explained by the fact that we took the native sequence from <i>Plasmodium falciparum </i>. The genome of <i>Plasmodium falciparum </i> is very A-T-rich ([http://areslab.ucsc.edu/cgi-bin/hgGateway UCSC Malaria Genome Browser]). |
| - | We think that expression of p25 and p28 may be improved by codon optimizing it for expression in bacteria like E. | + | We think that expression of p25 and p28 may be improved by codon optimizing it for expression in bacteria like ''E. coli'' and ''Asaia'' as we did with the immunotoxin. |
| - | Additional to the Western Blots, to rule out the possibility that concentration was too low, we | + | Additional to the Western Blots, to rule out the possibility that concentration was too low, we attempted a protein purification (following the [http://openwetware.org/wiki/Knight:Purification_of_His-tagged_proteins/Denaturing Knight protocol]) from a large culture volume (see figure) using Ni-NTA columns. |
Revision as of 18:59, 27 October 2010


Contents |
Proteins
We have chosen two different ways to target the parasite P.falciparum and prevent the malaria transmission through mosquitos. Our engineered bacteria could express either an immunotoxin, or two p-proteins, or even both for maximum efficiency. We tried to express all of these proteins using the C3 plasmid incorporating the strong promoter, a constitutive sequence for greater level of expression.
The Immunotoxin is one of our tools to block transmission of malaria parasite in mosquitos. It is composed of two main parts : The first one is a single-chain antibody fragment (scFv) directed to Pbs2l, which is a surface membrane protein of Plasmodium berghei . The second part is a lytic peptide, Shiva-1, which acts by forming “pores” on the parasite’s membrane. The immunotoxin is supposed to specifically target and lyse the parasite.
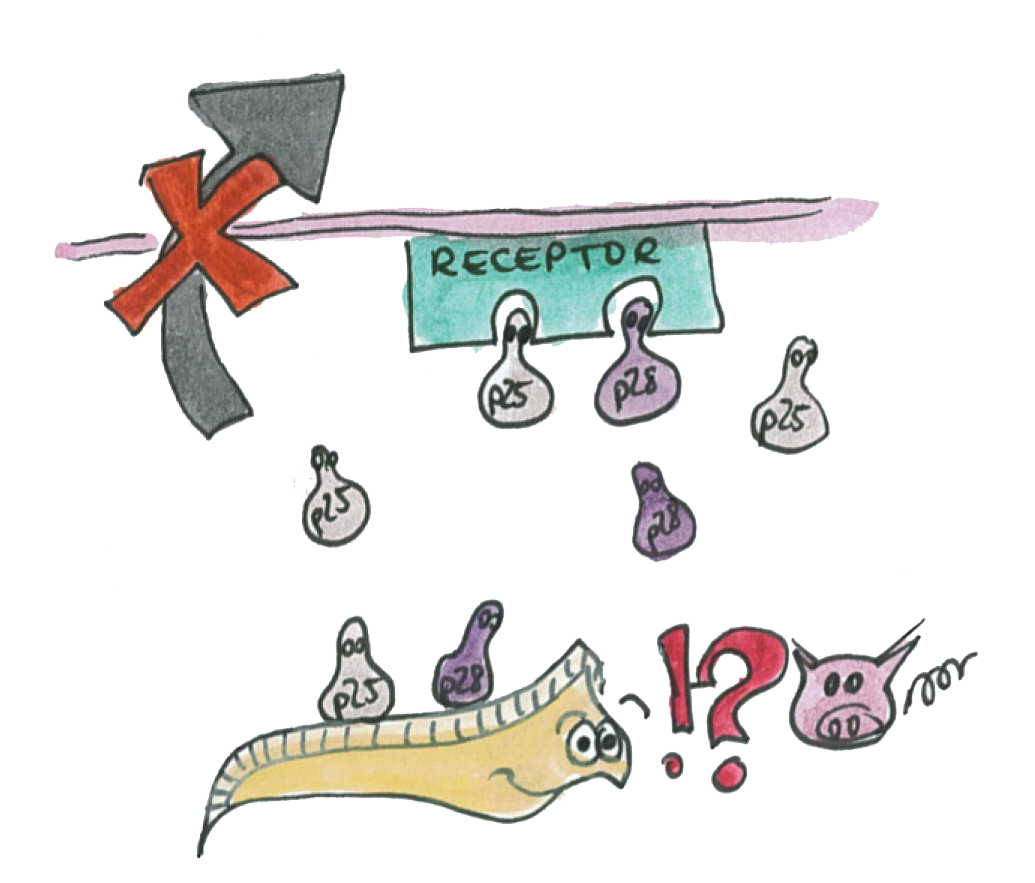
In parallel to the immunotoxin we thought of a different way of blocking the P.falciparum by using a group of proteins, called the "p-proteins".
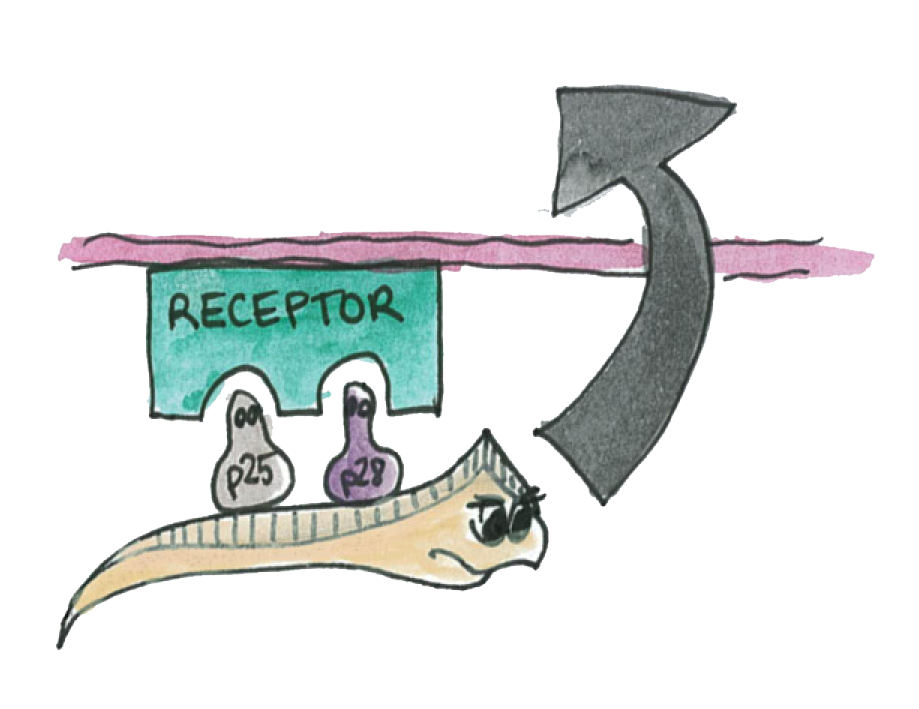
P25 and P28 are a class of important proteins expressed on the membrane of different type of Plasmodium; we call this ensemble of evolutionary conserved proteins the P-proteins. They are mainly expressed on the mosquito-stage parasite (ookinete). The ookinete has been intensively studied by scientists, looking for an ideal transmission-blocking vaccine target.
The Plasmodium normally uses its P-protein to interact with the epithelium in order to go through it.
Now if Asaia starts to produce a hight amout of these proteins, the interactions will be disrupted and the plasmodium will not propagate through the mosquito's epithelium.
Results
A: The Immunotoxin is expressed and appears in the supernatant
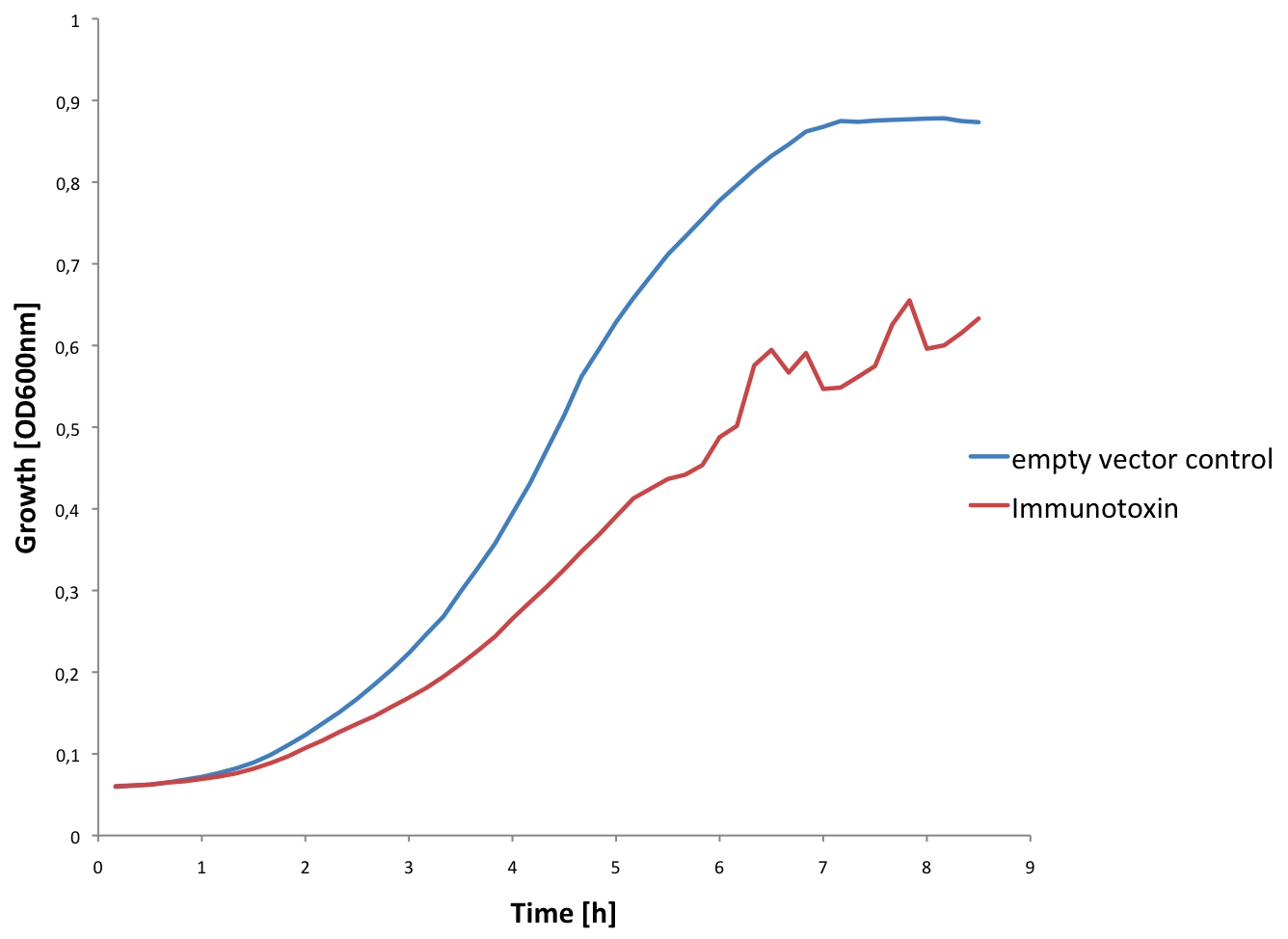
We tested expression of the immunotoxin in E. coli (see Materials and Methods for details). In a western blot analysis of whole cell lysates we could see bands corresponding to full length immunotoxin and possibly degraded fragments of the protein (see figure). The immunotoxin contains a PelB sequence that targets it for secretion into the periplasm. We concentrated the supernatant of both the immunotoxin and a control culture by running it through a filtering device with a 5 kDa cut-off. Running a western blot with these samples (see figure) we verified that the immunotoxin was also found in the supernatant.
B: The P-proteins are not expressed
The same experiments were conducted for the p-proteins p25 and p28. No bands were detected on the western blots (see figure) which leads us to the conclusion that these proteins were only very weakly expressed or not at all. This might be explained by the fact that we took the native sequence from Plasmodium falciparum . The genome of Plasmodium falciparum is very A-T-rich ([http://areslab.ucsc.edu/cgi-bin/hgGateway UCSC Malaria Genome Browser]). We think that expression of p25 and p28 may be improved by codon optimizing it for expression in bacteria like E. coli and Asaia as we did with the immunotoxin. Additional to the Western Blots, to rule out the possibility that concentration was too low, we attempted a protein purification (following the [http://openwetware.org/wiki/Knight:Purification_of_His-tagged_proteins/Denaturing Knight protocol]) from a large culture volume (see figure) using Ni-NTA columns.

 "
"