Team:ETHZ Basel/Achievements
From 2010.igem.org
(→Achievements Overview) |
(→Achievements Overview) |
||
| Line 8: | Line 8: | ||
<div class="thumbcaption"><div class="magnify"><a href="http://www.youtube.com/watch?v=mulRvAVExSc&hd=1" class="external" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div><b>Video 1: This video shows the E. lemming in action.</b><br />The unprocessed microscope images are available <a href="https://2010.igem.org/Team:ETHZ_Basel/Achievements/OriginalImages">here</a>.</div></div></div> | <div class="thumbcaption"><div class="magnify"><a href="http://www.youtube.com/watch?v=mulRvAVExSc&hd=1" class="external" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div><b>Video 1: This video shows the E. lemming in action.</b><br />The unprocessed microscope images are available <a href="https://2010.igem.org/Team:ETHZ_Basel/Achievements/OriginalImages">here</a>.</div></div></div> | ||
</html> | </html> | ||
| - | <html><div class="thumb tright"><div class="thumbinner" style="width: | + | <html><div class="thumb tright"><div class="thumbinner" style="width:402px;"> |
| - | <iframe title="YouTube video player" class="youtube-player" type="text/html" width=" | + | <iframe title="YouTube video player" class="youtube-player" type="text/html" width="400" height="325" src="http://www.youtube.com/embed/NThlEict4f4?rel=0&hd=1" frameborder="0"></iframe> |
<div class="thumbcaption"><div class="magnify"><a href="http://www.youtube.com/watch?v=NThlEict4f4&hd=1" class="external" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div><b>Short movie of an E. coli cell switching between the tumbling and the straight swimming modes.</b>.</div></div></div></html> | <div class="thumbcaption"><div class="magnify"><a href="http://www.youtube.com/watch?v=NThlEict4f4&hd=1" class="external" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div><b>Short movie of an E. coli cell switching between the tumbling and the straight swimming modes.</b>.</div></div></div></html> | ||
* '''The E. lemming: '''Watch the '''[https://2010.igem.org/Team:ETHZ_Basel/Achievements/E_lemming smallest remotely controlled robot in action]''' at ETH Zurich! The implementation using a chimeric fusion of Archeal Receptor to Bacterial chemotactic transducer enabled us to generate the first E. lemming. You find here recorded videos of live images of the light controlled ''E. coli'' - [https://2010.igem.org/Team:ETHZ_Basel/Achievements/E_lemming '''The E. lemming'''] is alive! | * '''The E. lemming: '''Watch the '''[https://2010.igem.org/Team:ETHZ_Basel/Achievements/E_lemming smallest remotely controlled robot in action]''' at ETH Zurich! The implementation using a chimeric fusion of Archeal Receptor to Bacterial chemotactic transducer enabled us to generate the first E. lemming. You find here recorded videos of live images of the light controlled ''E. coli'' - [https://2010.igem.org/Team:ETHZ_Basel/Achievements/E_lemming '''The E. lemming'''] is alive! | ||
Revision as of 14:55, 27 October 2010
Achievements Overview
The unprocessed microscope images are available here.
- The E. lemming: Watch the smallest remotely controlled robot in action at ETH Zurich! The implementation using a chimeric fusion of Archeal Receptor to Bacterial chemotactic transducer enabled us to generate the first E. lemming. You find here recorded videos of live images of the light controlled E. coli - The E. lemming is alive!
- BioBrick Toolbox: Get a glimpse of our collection of BioBricks that showcases the proteins we made not only for E. lemming's chemotaxis signaling network and cellular anchoring, but also for its light sensing & fluorescence reporting pathway, including our favorites: the light-sensitive couple PhyB-Pif3 and the Archeal light receptor-an intra-species fusion protein! You also find here the characterization of our parts.
- MATLAB Toolbox (Lemming Toolbox)
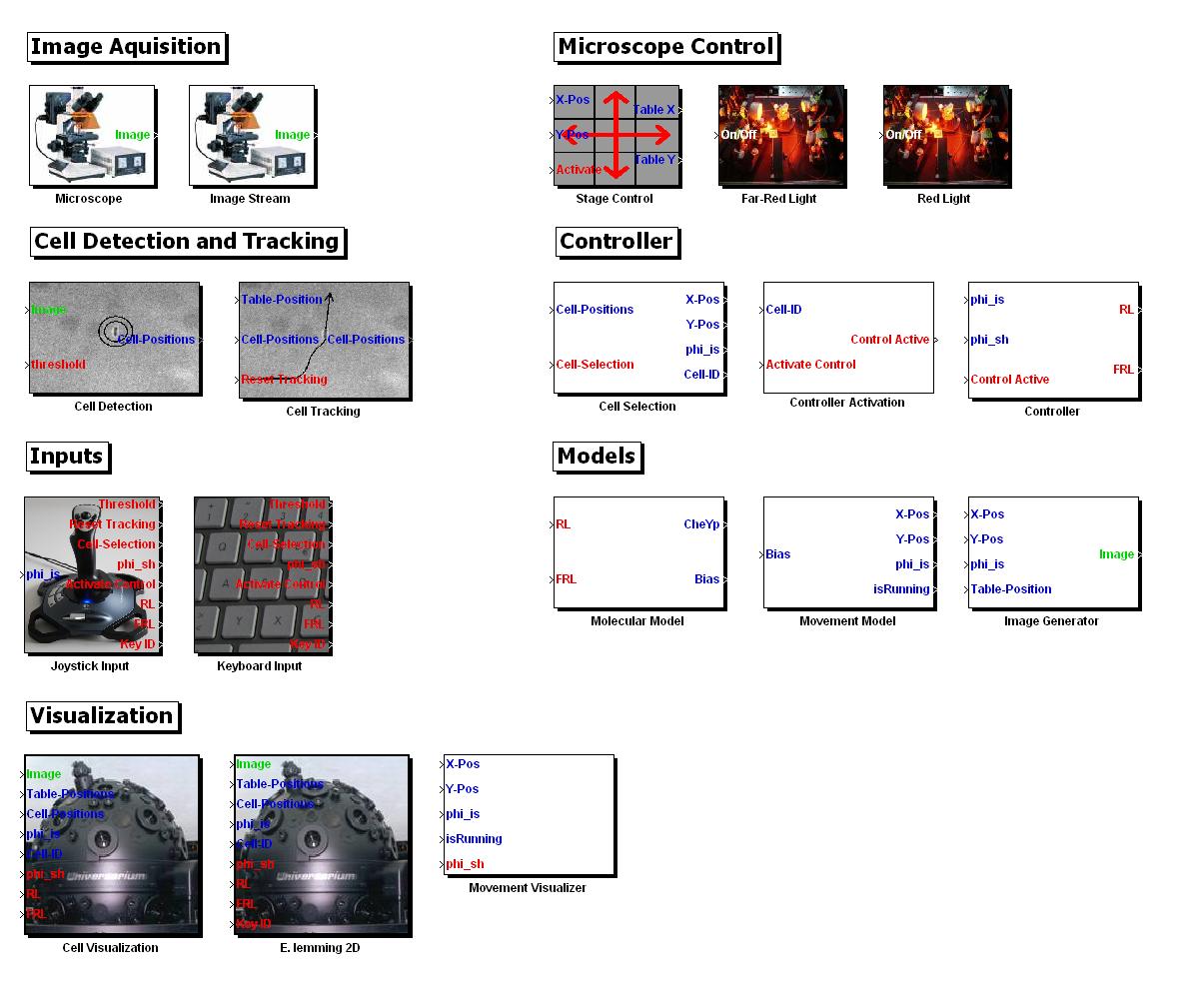
We managed to develop all our computer based tools & algorithms as re-usable, publicly available modules and we assembled the entire in silico setup of the E. Lemming into a MATLAB Toolbox! Our Toolbox includes 18 Simulink blocks encapsulating the algorithms we developed/implemented. We want to share our work with the iGEM community and interested persons and made it freely available for [http://sourceforge.net/projects/ethzigem10/files/LemmingToolbox_Setup.zip/download downloading].
- New Technical Standard
After weeks of applying the cloning strategy [http://dspace.mit.edu/handle/1721.1/46721 BBF RFC28], we came up with our own strategy on how BFF RFC28 could be made compatible with Tom Knight`s original assembly standard.
- Model of light-inducible chemotaxis pathway
We worked out a model to control chemotaxis by light. The idea was to instrumentalize light sensitive proteins (LSPs) in order to manipulate the concentrations of the chemotaxis proteins (eg CheY), through an intracellular anchoring reaction. As a consequence, we can take control on this pathway-and direct the E.lemming's movement!
Movement Model
We made a stochastic model to realistically simulate the chemotaxis motion of the cell in silico, which responds to the light input that is converted into changes in the chemotaxis pathway, that influence the motion of the E. lemming.
μPlateImager - software for the microscope
We developed μPlateImager, a platform independent software which enables the parallel acquisition of images and the remote control of light input signals. As a result, we achieved to control our E. lemmings with light, LIVE, while having them under the lens! Watch our short movie on this page!
E. lemming 2D - The Game
We invented a synthetic biology shooting game featuring our very own E. lemming! Give it a try and play with our E. lemming!
Controller
We developed five novel controllers for directing the E. lemming to the desired target. As stimuli for altering the bacterial movement, we use light inputs.
Systems Design
See how we managed to use our models to support the wetlab and vice versa when we designed the system.
 "
"


