Team:Edinburgh/Notebook/Red light sensor
From 2010.igem.org
Macbereska (Talk | contribs) (→Red Light Sensor) |
|||
| Line 485: | Line 485: | ||
<li>JM109 – RLS.lacZ</li> | <li>JM109 – RLS.lacZ</li> | ||
<li>JM109 - lacZ Control</li> | <li>JM109 - lacZ Control</li> | ||
| - | in LB + 40 Cml | + | in 2.5ml LB + 40 Cml |
| + | |||
| + | '''26/10/10''' | ||
| + | |||
| + | A modified version of the Beta-Galactosidase (Better Miller) Assay from OpenWetWare page http://openwetware.org/wiki/Beta-Galactosidase_Assay_(A_better_Miller) was used to attempt to characterise the RLS with lacZ. Aliquots were taken every hour of the cultures which were set up as before,100μl of ONs were used to inoculate two sterile LB + 40mg/ml Cml making up a total volume of 4ml in 5ml flasks which were then incubated in both light and dark, at 37C with shaking, as follows:</p> | ||
| + | |||
| + | <ul> | ||
| + | <li>JM109 – RLS.lacZ.YFP (Light/Dark)</li> | ||
| + | <li>envZ – RLS.lacZ.YFP (Light/Dark)</li> | ||
| + | <li>JM109 – RLS.lacZ (Light/Dark)</li> | ||
| + | <li>JM109 - lacZ Control (Light/Dark)</li> | ||
| + | </ul> | ||
| + | |||
| + | <p> For each sample at each of the time points, Abs420 / reaction time was calculated and plotted onto a graph, shown below. | ||
| + | |||
| + | |||
| + | [[Image:Ed10-RedLightSensCharData2.png]] | ||
Revision as of 18:19, 27 October 2010
Red Light Sensor
22/6/10
Transformation of BBa_M30109; well 14 H, distribution plate 3 (2010)--> red light sensor
24/6/10
Subcultured transformants from ampicillin plates to new plates
25/6/10
1) DNA mini-preps were prepared from the transformants 2) DNA mini-preps were cut with EcoRI enzyme to check for the presence of the plasmid (and DNA); agarose gel was prepared for DNA electrophoresis after cutting. 3) Results from cutting showed no presence of the sensor. Troubleshooting: was the plate number correct? Was procedure performed correct? by:Maria, Will, Marta
28/6/10
Tried to determine what was wrong with the red sensor that failed to appear in the gel. We definitely had the correct plate and well. We checked the seqeunce of the part and found that the sequence given does not match any given in BLAST. We suspect either that part of the plasmid was added to the distribution/registry or that the plasmid contains an illegal EcoR1 site, but are re-transforming cells just in case there was an experimental error. Due to failure of the previous transformation competent cells were retransformed with the two plasmids, as before. Two Chloramphenicol spread plates for each plasmid were made one of each pair containing 100micro-l of cells the other containing the 900micro-l remainder spun down and resuspended in 100micro-l of sterile LB. by: Richard, Lu
29/6/10
Streaking the colonies from the chloramphenicol plates on the new agar + chloramphenicol plates.
30/6/10
No growth on plates.
5/7/10
Made kanomycin 32mg/ml, IPTG 90mg/ml plates. Transformed cells with individual red light sensor parts (BBa_I15008 - 2010 distribution Plate 2 Well 13J; BBa-I15009 - distribution plate 1 Well 20F; BBa_I15010 - distribution Plate 3 Well 20D; & K098010 - distribution Plate 3 11N). Plated cells onto kanomycin plates and left to incubate overnight. by: Hannah and Richard
6/7/10
Richard/Hannah - made subcultures of red light sensor part transformants.
- Agarose Gel Eleckrophoresis of composite red sensor M30109
- The bands are not the right size
- Sequencing done by the registry starts in the middle of the sequence (for forward sequence)
- So it seems the plasmid doesn’t contain the right sequence
- We will try to assemble it from individual parts
8/7/10
Plasmid minipreps of I15009 (the only red sensor part transformant that grew in liquid culture) by: Richard and Hannah
9/7/10
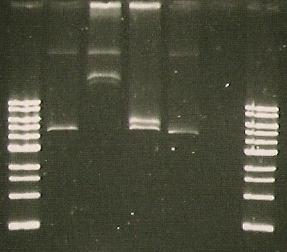
Gel run with I15009.
| Lane 1 | Lane 2 | Lane 3 | Lane 4 | Lane 5 | Lane 6 | Lane 7 | Lane 8 |
| Ladder | Pure SacB | Pure SacB & EcoRI | sacB & catR PCR | I15009 digest | S248T PCR Product | Purified 356K | Purified 356R |
- Insert Picture of Gel*
13/7/2010 NOTHING WORKS. No idea why...
16/7/10
Because the red light sensing parts were transformed with 50 µL or water instead of 15 µL during the first transformation, another attempted was tried. Transforming:
- I15008 (all red light sensor parts)
- I15009
- I15010
used 200 µL of cells, 800 µL of broth. Kan resistance.
23/07/10
Double digest of K098010. Ran gel. Lane was empty:
- Not going to bother saying what is in each lane.
30/7/10
Primers were ordered in order to PCR out the Red Light Sensor Parts from the [http://partsregistry.org/wiki/index.php?title=Part:BBa_M30109 Bio Brick BBa_M30109]
Forward Primer for PBad promoter [http://partsregistry.org/wiki/index.php?title=Part:BBa_I13453 Bio Brick BBa_I13453]
Reverse Primer for Heme Oxygenase (ho1) [http://partsregistry.org/wiki/index.php?title=Part:BBa_I15008 Bio Brick BBa_I15008]
Reverse Primer for Phycocyanobilin:Ferredoxin Oxidoreductase (PcyA) [http://partsregistry.org/wiki/index.php?title=Part:BBa_I15009 Bio Brick BBa_I15009]
Forward Primer for TetR repressible promoter [http://partsregistry.org/wiki/index.php?title=Part:BBa_R0040 Bio Brick BBa_R0040]
Reverse Primer for cph8 (Cph1/EnvZ fusion)Chimeric Cph1 light receptor/EnvZ protein [http://partsregistry.org/wiki/index.php?title=Part:BBa_I15010 Bio Brick BBa_I15010]
3/8/10
A PCR was run with the above primers in 3 distinct mixtures in an attempt to amplify out the Red Light Sensor parts using Bio Brick BBa_M30109 as a template.
PCR 1 Contained The PBad promoter Forward Primer & the PcyA Reverse Primer
PCR 2 Contained The TetR repressible promoter Forward Primer & the cph8 Reverse Primer
PCR 3 Contained The PBad promoter Forward Primer & the cph8 Reverse Primer
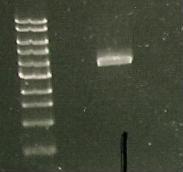
And a gel was run of the PCR products: Marker(Lane 1&6), only visible product- suggested PCR I15010 with RBS & promoter (Lane 3).

Due to the successful amplification of the I15010 cph8 Red light sensor part, the DNA was ligated into plasmid psb1C3.
4/8/10
Competent cells were transformed with the ligated psb1C3 plasmid containing I15010 and plated out on Cml 40 + IPTG 90 plates and incubated O/N at 37 degrees Celsius.
A PCR was run using the following primers:
PCR 1 Contained The BBa_B0034 RBS Forward Primer & the ho1 Reverse Primer
PCR 2 Contained The BBa_B0034 RBS Forward Primer & the PcyA Reverse Primer
PCR 3 Contained The PBad promoter Forward Primer & the ho1 Reverse Primer
PCR 4 Contained The PBad promoter Forward Primer & the PcyA Reverse Primer
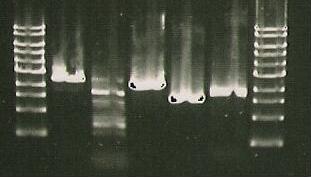
And a gel was run of the PCR products.
- Insert picture of Gel*
No successful amplification was seen.
11/8/10
Update from Maria. The transformations and PCR amplification of the HO1-pcyA fusion construct have nearly all failed. This weekend the kanamycin resistant version of the construct (K089010 in pSB3K3) was transformed and cultures grew. We already have a PCR product of the fused cph8-envZ sensing part. We now need to put the newly transformed K089010 in front of the sensor in order to rebuild the original sensor, which is well and truly fucked.
16/8/10
Successful PCR of K098010 using primers for EcoRI & I15009 reverse primer.
17/8/10
- The PCR product of the Harvard K098010 part was purified and then digested with both XbaI & SpeI as was the PCR product of cph8.
- These digestions were then ligated together ON using in two orientations.
18 &19/8/2010
- Fusion PCR C + H with R004081 & I15009-1
- also running H + C same primers as negative control
- run gel of PCR prod & 5 ul of ligation
- luxAB -SpeI; lumP -XbaI, ;uxCDE -XbaI
- lumP = P1024
- luxAB + lumP and luxAB + lyxCDE ligation
20/8/2010
- Fusion PCR with ?????
- get:
24/8/2010
- Digest cph8 (XbaI- yello) & Harvard (SpeI -brown)
- Ligate Harvard and cph8 = harvard + cph*8
- mutagenic PCR of lux CDE to mutate out site in LuxD
- subculture luxAB lumP transformants
- miniprep ON'S (?) set up
25/8/2010
- Minipreps of subcultures luxAB lumP,
- digest & run gel
- Purify luxCDE PCR product- from gel and froem the product; run the gel of purifications --> do MABEL self ligation- set 16C ON
- Fusion PCR harv + cph8- run gel of product
26/8/2010
- Gel1 - minipreps 1-5, gluxCDE, PluxCDE
- Gel2 - Minipreps 6-10, H + C ligation
- Lane 7: Harvard + Cph8 ligation
27/8/2010
Transform cells with self ligated MABEL mutated luxCDE from gel and PCR product
30/8/2010
- Lu wants a broad sward (?).
- Re-transform MABEL mutated
- luxCDE from gel- PCR products
- luxAB and luxABlumP double digest with EcoRi/PstI
- Fusion PCR if H + cph8 using H promoter forward and cph8 reverse primer
31/8/2010
- Lane 1- marker, Lane 2- H+ Cph8 fPCR
- ligated H+ C ON in psB1C3
- Double digest luxCDE with EcoRI and PstI - run gel
7/9/2010
- Run digest luxCDE with XbaI --> gel
- Gel of H + C ligation
- transform cells with
8/9/2010
- PCR with biobrick pre/suff - primers luxCDE 1 and 4
9/9/2010
- Purify luxCDE PCR
- Subculture transformants H + C
10/9/2010
- Add promoter to luxAB and luxABlumP
- Liquid ON'S of H + C subculture transformants
- Lab meeting: small-->
- sacB - some preeliminary sucrose sensitivity characterisation data.
- promoter: luxAB, luxAb-lumP- eission spectra, testing thing
- red light luciferase: S184T, 356 K
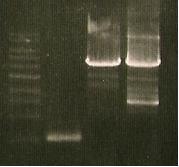
| Lane 1 | Lane 2 | Lane 3 | Lane 4 |
| Ladder | Tna up | Cph8- fixing prefix | Cph8 - fixing PstI site |
13/9/2010
- H + C + pSB1C3 minipreps
14/9/2010
- Digest H+ C minipreps with EcoRi. 100: 1,4,5. 900: 3,4,6
??? 20/9/2010
- Test luxCDE
- luxCDE digest with XBaI and PstI (buffer H)--> purify and ligate to luxAB digested with SPEI PstI in buffer B after purification.
- conclusion: purify digestions, ligate luxCDE into luxAB + promoter
21/9/2010
- Transform cells with luxAB + luxCDE ligation, plated on CML 40, IPTG 90
- Made up 50 ml liquid cultures of luxAB lumP and luxAB with Cml40, IPTG 90
04/10/10
Chris has managed to construct what we hope is a working red light sensor. It is being sent in for sequencing today.
Minor characterisation experiment:
-Red light sensor constructed with blue-white lacZ reporter
-If activated, the red light sensor transformants should produce blue colonies
-Red light sensor containing-colonies from transformant plate streaked onto two plates
-One plate left in the light, the other left in the dark
-If the one in the light grows blue and the one in the dark doesn't, it is a good indication that the construct is working
12/10/10
- Transform envZ mutant strain with YFP
- Plate out onto 40mg/ml Cml
13/10/10
- Subcultured envZ transformants onto Cml 40 plates
15/10/10
- Liquid Cultures made of transformants with Cml 40 & grown ON
16/10/10
- Liquid ONs viewed under blue light, yellow fluorescence seen showing successful transformation.
20/10/10
- RLS characterisation begins.
- Liquid ON's of
- JM109 – RLS.lacZ.YFP
- envZ – RLS.lacZ.YFP
- JM109 – YFP Control
- envZ – YFP Control
were made in 5ml LB + Cml 40 and grown at 37 degrees with shaking in the dark.
100μl of ONs were used to inoculate two sterile LB + 40mg/ml Cml making up a total volume of 4ml in 5ml flasks which were then incubated in both light and dark, at 37C with shaking, as follows:
- JM109 – RLS.lacZ.YFP (Light / Dark)
- envZ – RLS.lacZ.YFP (Light / Dark)
- JM109 – YFP Control (Light / Dark)
- envZ – YFP Control (Light / Dark)
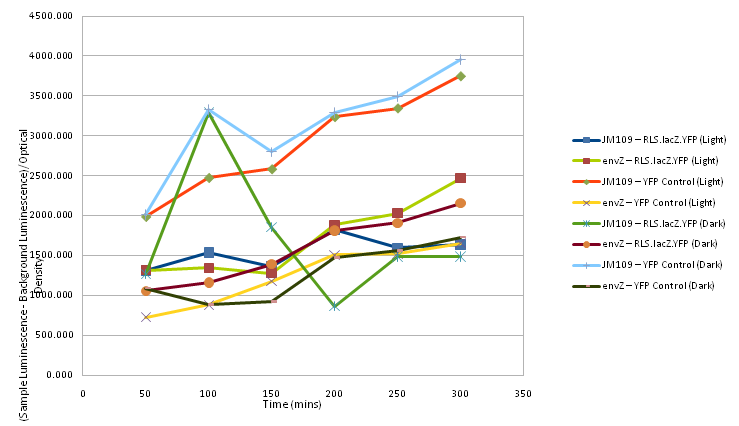
At 50 minute time intervals 200μl of each sample was taken and mixed with 800μl of sterile water in plastic cuvettes. The optical density of each sample was taken after the spectrophotometer was set with a sample of 200μl of sterile LB and 800μl water as a control, as was the luminescence, with readings for the background luminescence being taken for the sample of LB and water.
Two readings for each sample at each time interval were taken and then an average was calculated for each time interval for both optical density and luminescence.
For each sample at each time interval the average luminescence (normalised by the background luminescence) was divided by the optical density and plotted on a graph, shown below as Figure 2.
- PCR of PhoA promoter (from genomic DNA).
25/10/10
- Liquid ON's made up of
- JM109 – RLS.lacZ.YFP (Light/Dark)
- envZ – RLS.lacZ.YFP (Light/Dark)
- JM109 – RLS.lacZ (Light/Dark)
- JM109 - lacZ Control (Light/Dark)
For each sample at each of the time points, Abs420 / reaction time was calculated and plotted onto a graph, shown below. File:Ed10-RedLightSensCharData2.png
 "
"