Team:TU Delft/Notebook/cloning-plan
From 2010.igem.org
(→Cloning Plan) |
|||
| Line 5: | Line 5: | ||
Once we decided what we wanted to achieve and with which genes, we designed the following cloning plan. | Once we decided what we wanted to achieve and with which genes, we designed the following cloning plan. | ||
| - | At the start of our project we tried to perform [http://partsregistry.org/Assembly:3A_Assembly| three-way ligation] as described in the part registry. | + | At the start of our project we tried to perform [http://partsregistry.org/Assembly:3A_Assembly| three-way ligation] as described in the part registry. However, we experienced troubles by ligating small parts and therefore changed our approach to two-way ligation to make our BioBricks. |
==Two-way ligations strategies== | ==Two-way ligations strategies== | ||
| Line 54: | Line 54: | ||
==Our plan== | ==Our plan== | ||
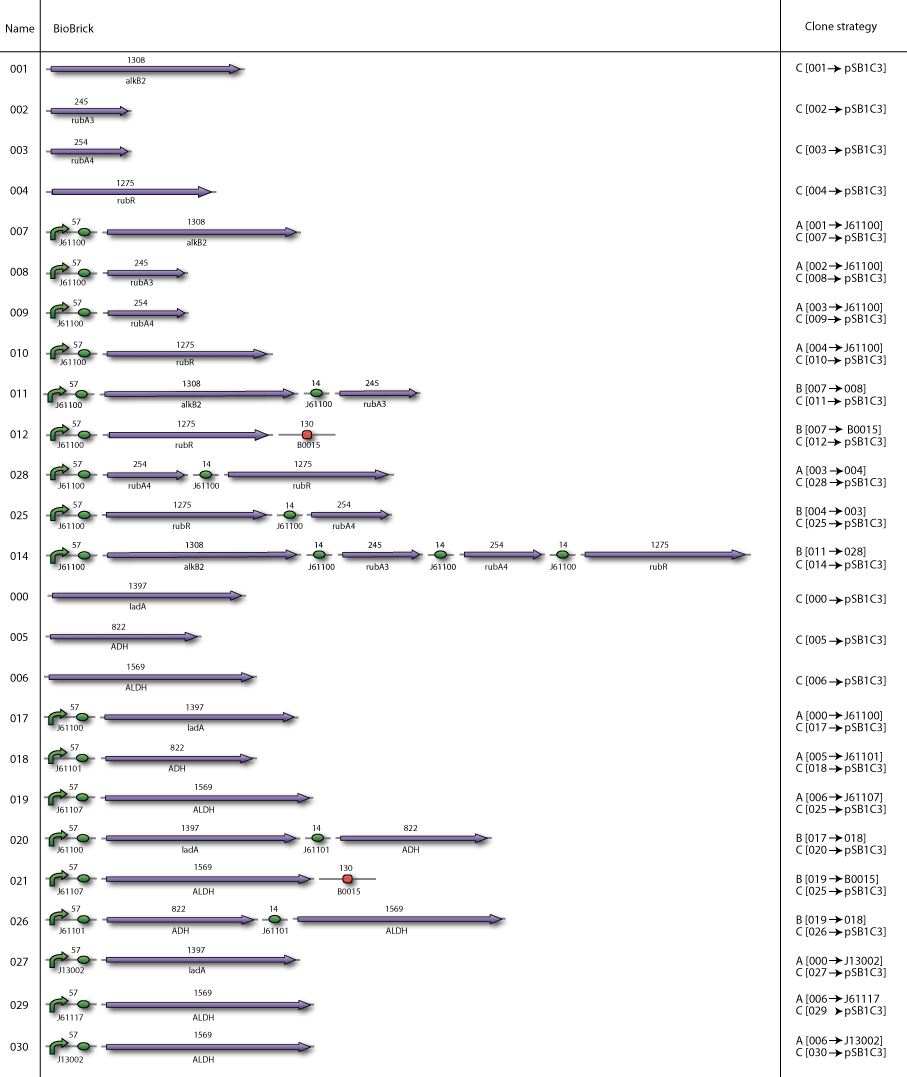
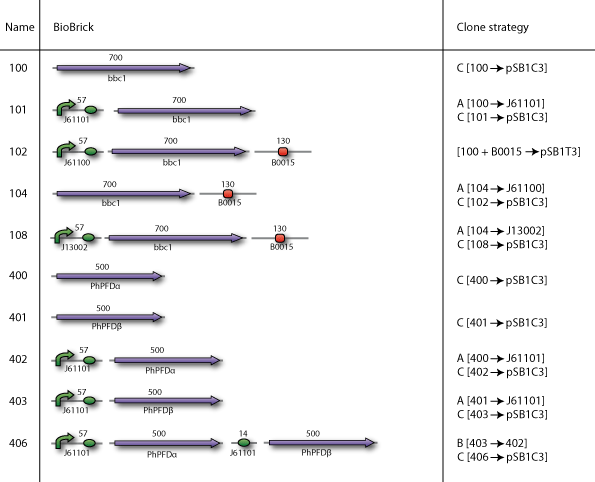
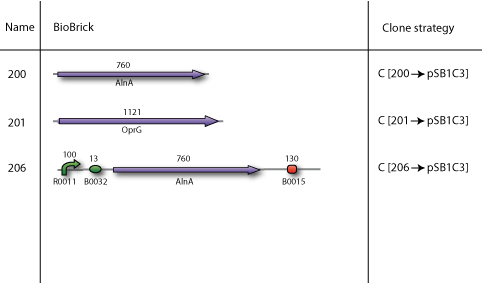
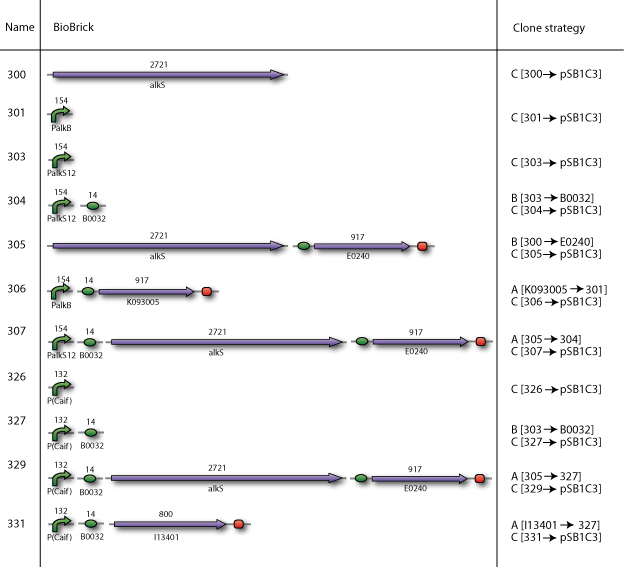
| - | Our BioBricks are | + | Our BioBricks are built from multiple parts. To make the final BioBrick several intermediates had to be prepared. Here our complete cloning plans of the different sub projects are illustrated. |
Revision as of 11:25, 27 October 2010
Cloning Plan
Once we decided what we wanted to achieve and with which genes, we designed the following cloning plan.
At the start of our project we tried to perform [http://partsregistry.org/Assembly:3A_Assembly| three-way ligation] as described in the part registry. However, we experienced troubles by ligating small parts and therefore changed our approach to two-way ligation to make our BioBricks.
Two-way ligations strategies
By using two-way ligation we ligated two parts to each other. The backbone of the smallest part was cut open. The largest part was cut out of its backbone. Then both parts were mixed and ligated. To prevent self-ligation we cut the backbone with an additional restriction enzyme.
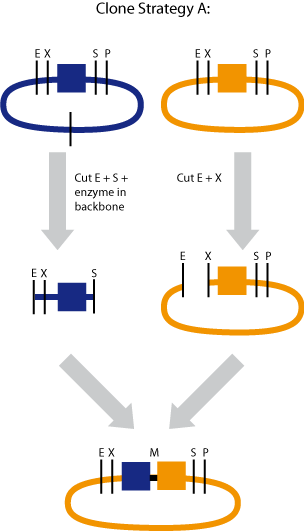
Cloning strategy A
In the case when the smallest part had to be downstream of the largest part, we used cloning strategy A
The smallest brick (blue) was cut with EcoRI and XbaI
The largest brick (orange) was cut with EcoRI and SpeI
It is important that the additional enzyme only cuts in the backbone and not in the part itself
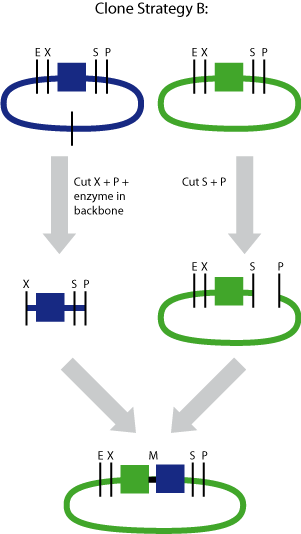
Cloning strategy B
In the case when the smallest part had to be upstream of the largest part, we used cloning strategy B
The smallest brick (green) was cut with SpeI and PstI
The largest part (blue) was cut with XbaI, PstI
It is important that the extra enzyme only cuts in the backbone and not in the part itself
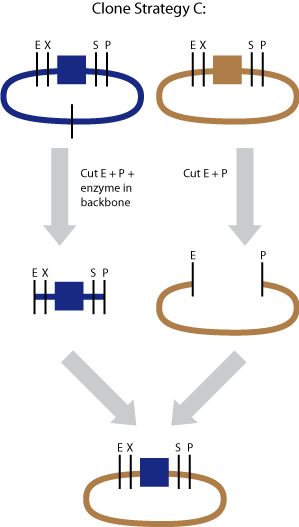
Cloning strategy C
In the end all BioBricks have to be cloned in the pSB1C3 plasmid backbone. With our strategy all the BioBricks are still in its original plasmid backbone, which in most cases was pSB1A2. We performed a pSB1C3 migration which is illustrated by cloning strategy C.
The part (blue) was cut with EcoI and PstI
To prevent self-ligation we cut the backbone with an additional restriction enzyme. It is important that the extra enzyme only cuts in the backbone and not in the part itself.
The vector backbone (brown) was cut with EcoRI, PstI
To prevent self-ligation we cut the insert with an additional restriction enzyme. In the case of the iGEM vectors is the insert J04450. It is important that the extra enzyme only cuts in the insert and not in the part itself.
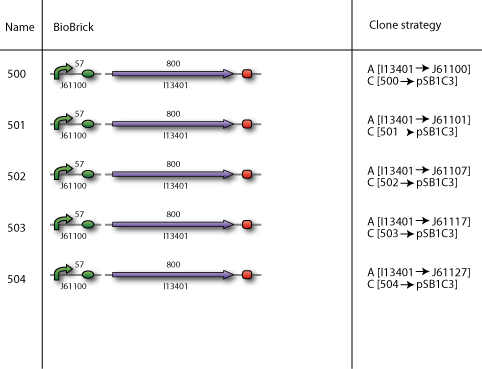
Our plan
Our BioBricks are built from multiple parts. To make the final BioBrick several intermediates had to be prepared. Here our complete cloning plans of the different sub projects are illustrated.
 "
"