Team:WashU/Notebook/Biobricks
From 2010.igem.org
(Difference between revisions)
(→2010/08/23) |
|||
| Line 123: | Line 123: | ||
5 H62 7.4 2.00 1.18 | 5 H62 7.4 2.00 1.18 | ||
6 H62 0.4 -0.65 0.17 | 6 H62 0.4 -0.65 0.17 | ||
| - | |||
| - | |||
*Preformed the following digestions at 37 degC for 2 hours and heat inactivation at 80 degC for 20 minutes: | *Preformed the following digestions at 37 degC for 2 hours and heat inactivation at 80 degC for 20 minutes: | ||
| Line 167: | Line 165: | ||
<center>[[Image:WashU_8-23Gel.jpg|400px]]</center> | <center>[[Image:WashU_8-23Gel.jpg|400px]]</center> | ||
<br> | <br> | ||
| + | |||
==2010/08/24== | ==2010/08/24== | ||
*The following transformations were analyzed | *The following transformations were analyzed | ||
Latest revision as of 07:26, 27 October 2010

2010/08/12
- Ordered the following primers to biobrick parts
p18 Forward KanMX4 ATT CTT AGA ATT CGC GGC CGC TTC TAG CCC GCC GCC ACC ATG GGT AAG GAA AAG ACT CAC GTT TCG p19 Reverse KanMX4 GCC GCC CTC TGC AGC GGC CGC TAC TAG TAT TAG AAA AAC TCA TCG AGC ATC AAA TGA AAC TG p20 Forward NatMX4 GTT TCT TCG AAT TCG CGG CCG CTT CTA GCC CGC CGC CAC CAT GGG TAC CAC TCT TGA CGA CAC G p21 Reverse NatMX4 GTT TCT TCC TGC AGC GGC CGC TAC TAG TAT TAG GGG CAG GGC ATG CTC ATG TAG p22 Forward Ura3-1 GCACAGAACAATAACCTGCTGGAAACGAAGATAAATCgaagacGATTACTTCGCGTTATGCAGGC p23 Reverse Ura3-1 GCATCTTCTCAAATATGCTTCCCAGCCTGCTTATCcttctgAAATTCTGCCTCGTGATACGCC p24 Forward Ura3-2 tactagtagcggccgctgcagTCTTAACCCAACTGCACAGAACAATAACCTGCTGGAAACG p25 Reverse Ura3-2 ctctagaagcggccgcgaattcTTAGTATTGCTGGCCGCATCTTCTCAAATATGCTTCCCAGCC p26 Forward His3-1 GAACAGGCCACACAATCGCAAGTGATTAACgaagacGATTACTTCGCGTTATGCAGGC p27 Reverse His3-1 CCTTGAACGCACTCTCACTACGGTGATGATCActtctgAAATTCTGCCTCGTGATACGCC p28 Forward His3-2 tactagtagcggccgctgcagAGAGGGAGAAGCAGTAGCAGAACAGGCCACACAATCGCAAG p29 Reverse His3-2 ctctagaagcggccgcgaattcTTATGGCAACCGCAAGAGCCTTGAACGCACTCTCACTACGG p30 Biobrick Forward tgccacctgacgtctaagaa p31 Biobrick Reverse attaccgcctttgagtgagc p32 Forward Ura3 Check CGG TAA TCT CCG AGC AGA AGG AAG AAC G p33 Reverse Ura3 Check CAT TAC GAC CGA GAT TCC CGG GTA ATA ACT G p34 Forward His3 Check GAG CAG AAA GCC CTA GTA AAG CGT ATT ACA AAT G p35 Reverse His3 Check CTA CAT AAG AAC ACC TTT GGT GGA GGG AAC p36 Forward SxL gtttcttcgaattcgcggccgcttctagagcccgccgccaccatgtacg p37 Reverse SxL gtttcttcctgcagcggccgctactagtattattataccttgcgctttttcttgggg p38 Vector internal Sequencing 1 - Forward 1400 start ACA CCC GTC CTG TGG ATC p39 Vector internal Sequencing 2 - Reverse 1523 start GAAGTGGCGAGCCCGATCTTC p40 Vector internal Sequencing 3 2301 start CCACCTCGACCTAACTCGAGTTAC
2010/08/19
- The following PCr reactions were run and then column Purified.
Number Product Template Forward Primer Reverse Primer Temperature 1 Kan BB KanMX4 p18 p19 58 2 Nat BB NatMX4 p20 p21 58 3 Kan BB KanMX4 p18 p19 60 4 Nat BB NatMX4 p20 p21 60 5 Kan BB KanMX4 p18 p19 64 6 Nat BB NatMX4 p20 p21 64
- The following digestions of were run.
K58 N58 K60 N60 K64 N64 P1 H20 12.5 12.5 12.5 12.5 12.5 12.5 22.5 Buffer 2 5 5 5 5 5 5 5 BSA 0.5 0.5 0.5 0.5 0.5 0.5 0.5 DNA 30 30 30 30 30 30 20 EcoRI 1 1 1 1 1 1 1 PstI 1 1 1 1 1 1 1
- The digestions were run for 6 hours at 27 degC and then kept at 4 degC o/n
- Made Chloramphenicol Plates at a concentration of 34μg/ml
2010/08/21
- The ligation reactions (KFYP3, KYFP4,KYFP5, - control, + control) were transformed and plated onto amp plates.
- Transformation Results
Plate Result 1. K58&p1 2 colonies formed, one on the edge of the plate pointing towards a possible satellite colony 2. N58&p1 No colonies appeared to have formed 3. K60&p1 No colonies appeared to have formed 4. N60&p1 No colonies appeared to have formed 5. K64 &p1 No colonies appeared to have formed 6. N64 & p1 No colonies appeared to have formed pSB1AT3 Many ~100 colonies formed with over 50% expressing red color under room light, predominantly large colonies are red pSB1C3 ~30 colonies formed with slght red coloring at the center
- Over Night Colonies prepared from
K58&p1 colony from middle of plate 34μg/ml chloramphenicol liquid media pSB1AT3 50μg/ml Amp liquid media pSB1C3 34μg/ml chloramphenicol liquid media
2010/08/22
- Miniprepped the following from overnight cultures made on 8/21/10:
DNA Concentration (ng/μl) 260/280 20/230 Kan BB in pSB1C3 37.8 1.92 2.15 pSB1AT3 with BBa_J04450 insert 102.1 1.92 2.26 pSB1C3 with BBa_J04450 insert 196.8 1.91 2.31
- The following PCR reaction was run with an elongation time of 3.5 minutes:
Number Product Template Forward Primer Reverse Primer Annealing Temperature 1 His3 Intermediate Vector pSB1AT3 w/ insert p27 p29 58 2 Ura3 intermediate Vector pSB1AT3 w/ insert p23 p25 58 3 His3 intermediate Vector pSB1AT3 w/ insert p27 p29 60 4 Ura3 intermediate Vector pSB1AT3 w/ insert p23 p25 60 5 His3 intermediate Vector pSB1AT3 w/ insert p27 p29 62 6 Ura3 intermediate Vector pSB1AT3 w/ insert p23 p25 62
- The following Digestion was run:
pSB1C3 w/ insert H20 40 DNA 2.5 NEB-3 5 BSA 0.5 EcoRI 1 PstI 1
- The following Gel was run:
1 1kb Ladder 2 H58 intermediate 3 U58 intermediate 4 H60 intermediate 5 U60 intermediate 6 H62 intermediate 7 U62 intermediate 8 pSB1C3 Digestion product
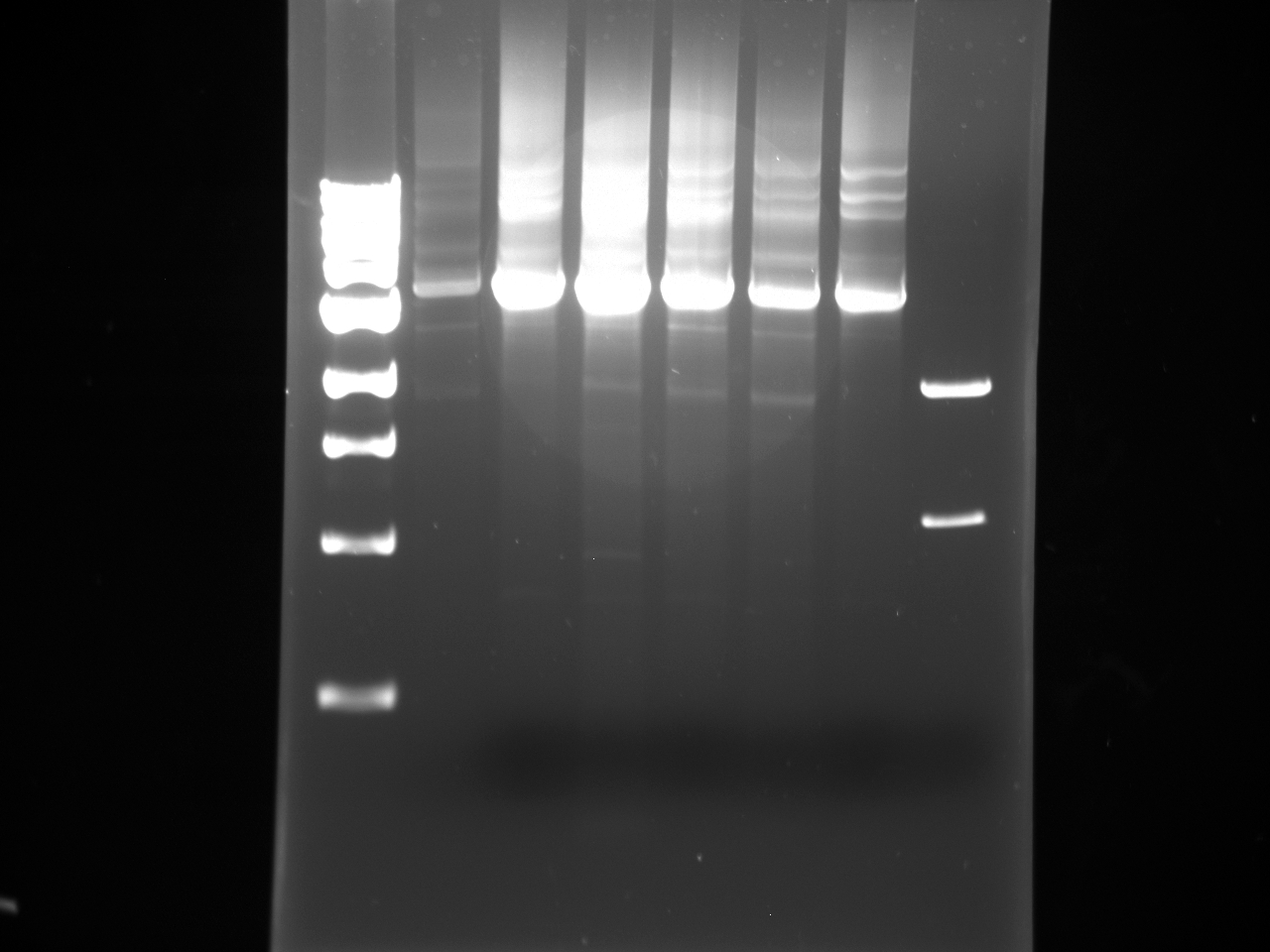
- The gel was cut and the products were gel purified using the Sigma Kit
- The following PCR reaction was let run over night with an elongation time of 3.5 minutes:
Number Product Template Forward Primer Reverse Primer Annealing Temperature 1 His3 Final Vector H58 Intermediate Vector p27 p29 58 2 Ura3 Final Vector U58 Intermediate Vector p23 p25 58 3 His3 Final Vector H60 Intermediate Vector p27 p29 60 4 Ura3 Final Vector U60 Intermediate Vector p23 p25 60 5 His3 Final Vector H62 Intermediate Vector p27 p29 62 6 Ura3 Final Vector U62 Intermediate Vector p23 p25 62
2010/08/23
- Nanodropped Over night PCR product:
Number Label Concentration ng/μL 260/280 260/230 1 H58 10.2 2.34 1.22 2 U58 6.6 2.31 1.13 3 H60 10.2 1.81 1.14 4 U60 5.5 2.33 1.16 5 H62 7.4 2.00 1.18 6 H62 0.4 -0.65 0.17
- Preformed the following digestions at 37 degC for 2 hours and heat inactivation at 80 degC for 20 minutes:
H58 U58 H60 U60 H62 pSB1C3 w/ insert ADH1 Promoter Kan_BB Linear pSB1T3 pSB1AT3 w/ insert H20 12.5 12.5 12.5 12.5 12.5 40 40 29.3 32.5 335.6 DNA 30 30 30 30 30 2.5 2.5 13.2 10 4.9 NEB-2 - - - - - - 5 5 5 5 NEB-3 5 5 5 5 5 5 - - - - BSA 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 1μL E1 EcoRI EcoRI EcoRI EcoRI EcoRI EcoRI EcoRI XbaI EcoRI EcoRI 1μL E2 PstI PstI PstI PstI PstI PstI SpeI PstI PstI PstI 1μL E3 XbaI 1μL E4 SpeI
- Preformed the following ligations following the Biobrick Manual Protocol
H58 U58 H60 U60 H62 AHD1+K_BB+pSB1T3 ADH1+K_BB+pSB1AT3 H20 11 11 11 11 11 11 11 Component 1 H58 U58 H60 U60 H62 ADH1 ADH1 Component 2 pSB1C3 pSB1C3 pSB1C3 pSB1C3 pSB1C3 K_BB K_BB Component 3 Linear pSB1T3 pSB1AT3 10x T4 ligase buffer 2 2 2 2 2 2 2 T4 DNA Ligase 1 1 1 1 1 1 1 1
- Preformed the following transformations:
1 H58 Amp 2 U58 Amp 3 H60 Amp 4 U60 Amp 5 H62 Amp 6 N58+pSB1C3 Chlor 7 N60+pSB1C3 Chlor 8 N64+pSB1C3 Chlor 9 ADH1+Kan_BB+pSB1T3 Tet 10 ADH1+Kan_BB+pSB1AT3 Amp 11 Tet - Control Tet 12 Chlor - Control Chlor
- Ran the following Gel:
1 2 3 4 5 6 7 8 1000bp Ladder H58 U58 H60 U60 H62 U62 1000bp
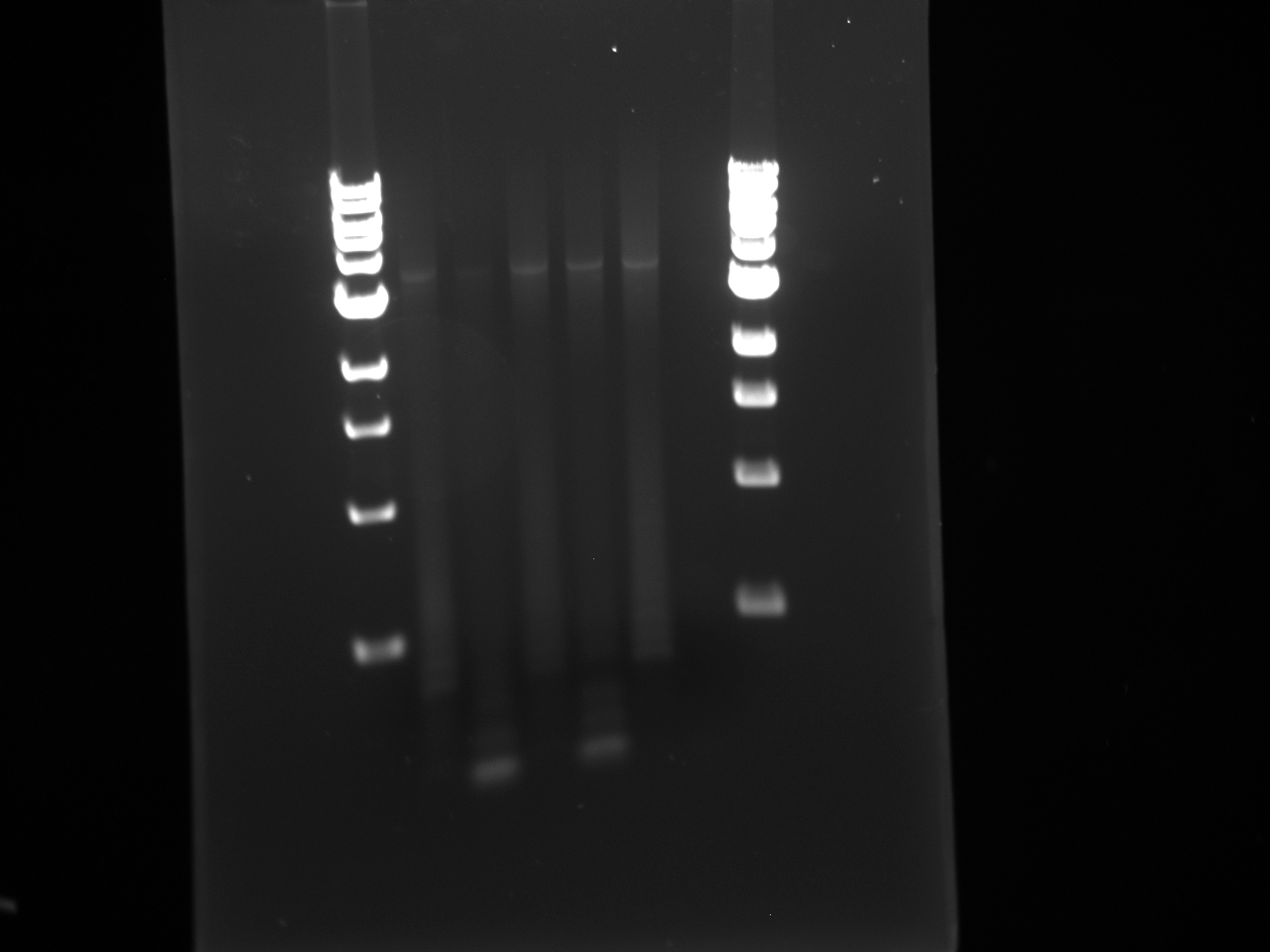
2010/08/24
- The following transformations were analyzed
1 H58 Amp No colonies were observed 2 U58 Amp No colonies were observed 3 H60 Amp No colonies were observed 4 U60 Amp No colonies were observed 5 H62 Amp No colonies were observed 6 N58+pSB1C3 Chlor No colonies were observed 7 N60+pSB1C3 Chlor No colonies were observed 8 N64+pSB1C3 Chlor No colonies were observed 9 ADH1+Kan_BB+pSB1T3 Tet A Lawn was observed 10 ADH1+Kan_BB+pSB1AT3 Amp ~ 80 colonies were observed with ~ 10% showing a red coloring 11 Tet - Control Tet A Lawn was observed 12 Chlor - Control Chlor No colonies were observed
- Conclusions:
- The tetracycline plates did not succesfully select for resistance
- The following PCR reactions were run using the sigma supplied PCR kit:
H64 U64 H68 U68 C68 Initial Tm 64 64 64 64 64 Second Tm 64 64 68 68 68 H20 37.5 37.5 37.5 37.5 42.5 PCR Buffer 5 5 5 5 5 2.5μL Primer 1 p26 p22 p26 p22 None 2.5μL Primer 2 p27 p23 p27 p23 None pSB1AT3 10ng/μL1 1 1 1 1 Nucleotide Mix 1 1 1 1 1 Taq Polymerase 0.5 0.5 0.5 0.5 0.5
- These reagents were made following the protocol located at ??
- The following PCR program was used
1. 94 degC 2:00 2. 94 degC 0:30 3. Initial Tm 1:00 4. 72 degC 3:30 Go to Step 2 x5 5. 94 degC 0:30 6. Second Tm 1:00 7. 72 degC 3:30 Go to Step 5 x 30 8. 68 degC 10:00 9. 4 degC overnight
- The following digestion was run and Gel Purified:
pSB1AT3 x3 pSB1C3 x3 H20 37.5 40 DNA 5 (102.1ng/μL) 2.5 (197ng/μL) NEB-3 5 5 BSA 0.5 0.5 EcoRI 1 1 PstI 1 1
- Digestion Program:
1. 37 degC 2:00:00 2. 80 degC 20:00
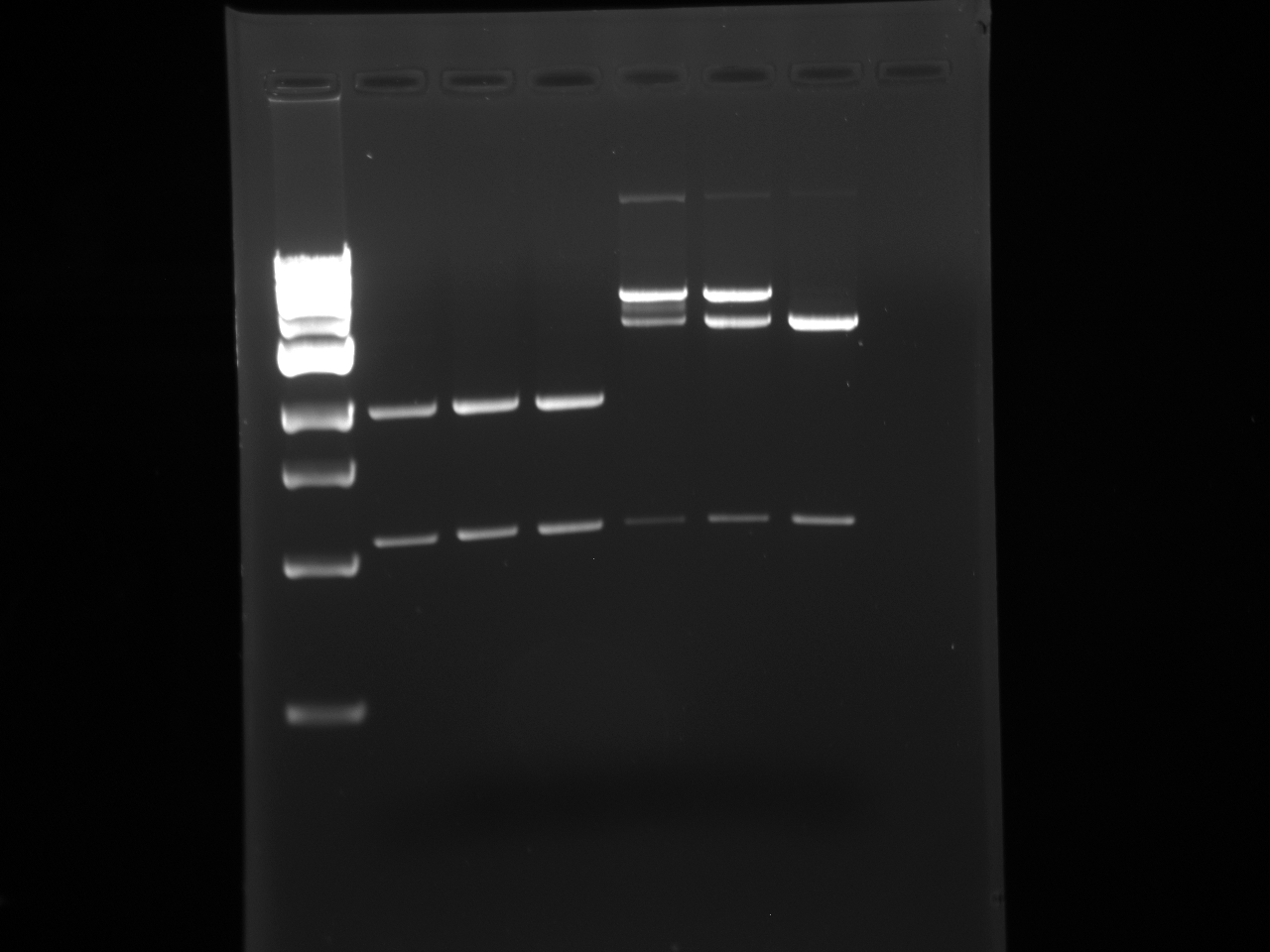
1 2 3 4 5 6 7 1kb ladder pSB1C3 pSB1C3 pSB1C3 pSB1AT3 pSB1AT3 pSB1AT3
- The second shortest band representing the linearized plasmid backbone was cut from each column and gel purified.
- Gel purification was conducted using the Sigma kit, the the elution solution was heated to 65 degrees to accommodate the large Linear DNA fragment.
- Lanes 2&3 and 4&5 were combined on one column to increase concentration.
- All elutions were done with 30 μL in order to increase concentration
- Nanodrop results:
- linear pSB1C3 from lane 1: 9.1 ng/uL
- linear concentrated pSB1C3 from lanes 2&3: 8.1 ng/ul
- linear pSB1AT3 from lane 4: 4.1 ng/ul
- linear concentrated pSB1AT3 from lanes 5&6: 5.5 ng/ul
- New LB and Amp Plates were Made
- The Kan_BB was submitted to sequencing
- 1μM dilutions of the biobrick primers were made.
- The following sequencing reaction was set up for both the forward and reverse primers:
5.3 μL of 37ng/μL Kan_BB DNA 3.2 μL of 1μM plasmid 3.5 μL of H20
2010/08/25
- Nanodropped the products of the PCR-1 on 8/24 of the Ura3 and His3 vectors
- A blank that underwent the PCR reaction with primers added only after the reaction was run to completion was used.
ng/μL 260/280 260/230 U64 570.1 1.72 1.41 H64 594.6 1.80 1.59 U68 585.3 1.79 1.61 H68 350.9 1.71 1.57
- The PCR-1 Products were gel Purified
1 1kb Ladder 2 Ura3 at 64, 64 degC No PCR Product Formed 3 His3 at 64, 64 degC Excised ~3.5kb band 4 Ura3 at 64, 68 degC No PCR Product Formed 5 His3 at 64, 68 degC Excised ~3.5kb band 6 Control at 64, 68 degC The 10ng of template doesn't show up on the gel
- Lanes 3 and 5 were gel purified with the sigma kit, in 25 μL elution that was preheated to 65 degC.
ng/μL 260/280 260/230 H64 16.2 2.35 0.11 H68 32.2 1.93 0.06
- 12 Kanamycin and 12 tetracycline plates were made
- The Following Tranformations were conducted:
- pSB1AT3 was obtained from miniprepped freezer stock, pSB1K3 was obtained from a iGEM well plate
Number Plasmid Plate 1 pSB1AT3 Tet 2 None Tet 3 pSB1AT3 Amp 4 None Amp 5 pSB1K3 Kan 6 None Kan
- The Following PCR reactions were let run overnight:
1 2 3 4 5 6 7 8 U64-1 H64-2 U68-1 H68-2 K64 N64 K60 N71 Initial Tm 64 64 64 64 64 64 60 68 Second Tm 64 64 68 68 64 64 64 71 H20 37.5 37.5 37.5 37.5 37.5 37.5 37.5 37.5 PCR Buffer 5 5 5 5 5 5 5 5 2.5μL Primer 1 p22 p28 p22 p28 p18 p20 p18 p20 2.5μL Primer 2 p23 p29 p23 p29 p19 p21 p19 p21
pSB1AT3 H64-1 pSB1AT3 H68-1 KanMX4 NatMX4 KanMX4 NatMX4
Nucleotide Mix 1 1 1 1 1 1 1 1 Taq Polymerase 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 Annealing Time 3:30 3:30 3:30 3:30 3:30 3:30 1:00 1:00
- These reagents were made following the protocol located at ??
- The following PCR program was used
1. 94 degC 2:00 2. 94 degC 0:30 3. Initial Tm 1:00 4. 72 degC 3:30 Go to Step 2 x5 5. 94 degC 0:30 6. Second Tm 1:00 7. 72 degC 3:30 Go to Step 5 x 30 8. 68 degC 10:00 9. 4 degC overnight
3020/08/30
- Ligations from 8/26 were transformed into E. coli:
Number Transformation Resistance 1 Negative Amp control Ampicillin 2 Negative Chloramphenicol controlChloramphenicol 3 His-64_2 with RFP Ampicillin 4 His-68_2 with RFP Ampicillin 5 Kan64 in pSB1C3 Chloramphenicol 6 Nat64 in pSB1C3 Chloramphenicol 7 Kan60 in pSB1C3 Chloramphenicol 8 Nat71 in pSB1C3 Chloramphenicol 9 kYFP3 in pSB1C3 Chloramphenicol
- Made new LB Amp plates
2010/09/02
- Analyzed results of transformation from 8/30
Number Transformation Resistance Results 1 Negative Amp control Ampicillin No colonies 2 Negative Chloramphenicol control Chloramphenicol No colonies 3 His-64_2 with RFP Ampicillin Three groups of ~ 150 colonies, no red colonies 4 His-68_2 with RFP Ampicillin 3 colonies- 1 large red one 5 Kan64 in pSB1C3 Chloramphenicol 12 colonies 6 Nat64 in pSB1C3 Chloramphenicol ~70 colonies 7 Kan60 in pSB1C3 Chloramphenicol 14 colonies 8 Nat71 in pSB1C3 Chloramphenicol 6 colonies- 1 red coloniy 9 kYFP3 in pSB1C3 Chloramphenicol lawn of red colonies
2010/09/06
- Ran the following Colony PCR Reactions and plated the colonies onto a plate.
1. K60-1 2. K60-2 3. K60-3 4. K60-4 5. K60-5 6. K64-1 7. K64-2 8. K64-3 9. K64-4 10. K64-5 11. N64-1 12. N64-1 13. N64-3 14. N64-4 15. N64-5 16. N71-1 17. N71-2 18. N72-3 19. N72-4 20. N72-5 21. SxL_BB-1 22. SxL_BB-1
- Reactions 1-20 were run using the following PCR mix form the Sigma Kit:
37.5 μL H20 5μL PCR Buffer 1μL DNucleotides 0.5μL Taq polymerase 2.5μL p30 2.5μL p31
- Reactions 21 and 22 were run using the following PCR mix from the Sigma Kit:
37.5 μL H20 5μL PCR Buffer 1μL DNucleotides 0.5μL Taq polymerase 2.5μL p36 2.5μL p37
- All of the PCR reactions were run with annealing temperatures of 67 ° C.
- The PCR reactions were then loaded onto a gel in the following order with 100bp ladders
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 Lad. 1 2 3 4 5 6 Lad. 7 8 9 10 11 12 Lad
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 Lad 13 14 15 16 16 17 Lad 18 20 21 22 Lad
- PCR reaction 16 was accidently loaded onto the gel twice. Lower lanes 7 and 8 may be shifted to contain PCR mix 19 instead of 17 or 18, so they should be treated with suspicious.
- Expected Results:
- Expected Results =gene length +316 for the flanking regions between primers and insert
- Lanes U2-U12 will run at 810+316=1126
- Lanes U13-L10 will run at 573+316=889
- Lanes U11, U12 will run at 1000+316=1316
- The gels were observed using a handheld UV light and 4 different banding patterns were observed.
- Pattern A: No Banding observed
- Pattern B: A band right below the second highest band (1,200bp)
- Pattern C: Well flouresced
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 Lad A B A B B A B Lad A A B A A Lad
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 Lad A C Smear A A A A Lad A C C A Lad
2010/09/10
- The colonies plated from the colony PCR on monday were mistakenly plated onto an Amp plate so no growth or viable colonies were observed.
- The following colony PCR was run.
1. K60-1 2. K60-2 3. K60-3 4. K60-4 5. K60-5 6. N64-1 7. N64-2 8. N64-3 9. N64-4 10. N64-5 11. N71-1 12. N71-2 13. N71-3 14. N71-4 15. N71-5
- The following PCR master mix was made using the Sigma kit then used with reaction volumes of 30μL.
392.7μL H20 51μL Buffer 10.2μL dNTP 25.5μL p30 25.5μL p31 5.1μL Taq
2010/09/11
- A PCR was run on U64-1 and U68-1 Using p24, p25 and the PCR Master Mix that was previously made from the sigma kit. *The following program was used
1. 1:00 94 degC 2. 0:30 94 degC 3. 1:00 64 degC 4. 3:30 72 degC GoTo 2 4X 5. 0:30 94 degC 6. 3:30 68 degC GoTo 5 29X 7. 10:00 72 degC 8. Forever 4 degC
- A Gel was run with the Colony PCR samples from 9/10.
- The gel was made with 8ml 5XTBE in 40 ml Water and 0.4g Agarose and 0.8μL EtBr
- 10μL were loaded into each lane and a 100bp Ladder was used.
- The lanes of the gel are as follows
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 Lad K60-1 K60-2 K60-3 K60-4 K60-5 N64-1 N64-2 N64-3 N64-4 N71-1 N71-2 N71-3 N71-4 Lad
- Expected Results:
- Expected Results =gene length +316 for the flanking regions between primers and insert
- Lanes 2-6 will run at 810+316=1126
- Lanes 7-14 will run at 573+316=889
- Results:
1. Ladder 2. 11,000bp fragment, birght 3. 11,000bp fragment 4. 11,000bp fragment bright 5. 300bp fragment 6. 300bp fragment 7. 550bp fragment 8. 400bp fragment 9. 500bp fragment 10. 13,000-14,000bp fragment, faint 11. 200bp fragment 12. 200bp fragment 13. 200bp fragment 14. 200bp fragment 15. Ladder
- Overnight Cultures were made of lanes 2 and 10 which corresponded to K60-1 and N64-4
2010/09/16
- A PCR was run on SxL in order to create SxL_BB
- A stock solution of the SxL construct sent by Mr. Gene was made a 10ng/µL
- The PCR was made using 44µL of the master mix made from the sigma kit
- 2.5 µL of p36 and p37 were used
- The following program was let run
1. 94 degC 3:00 2. 94 degC 0:30 3. 62 degC 0:30 4. 72 degC 1:00 GOTO 2 x5 5. 94 degC 0:30 6. 68 degC 1:30 GOTO 5 x30 7. 72 degC 10:00 8. 4 degC
2010/09/18
- A PCR was run on NatMX4 in order to make Nat_BB
- The following reagents were used
44µL PCR master mix made from sigma ket 2.5µL 10µM p20 2.5µL 10µM p21 1µL 10ng/µL NatMX4
- The following conditions were used:
1. 94 degC 3:00 2. 94 degC 0:30 3. 66 degC 0:30 4. 72 degC 1:00 GOTO 2 x5 5. 94 degC 0:30 6. 71 degC 1:15 GOTO 5 x30 7. 72 degC 10:00 8. 4 degC forever
- A Colony PCR on H_Vector was run
- p30 and p31 primers are expected to give a band of 1000bp if RFP was inserted and 330 if nothing was inserted
- p30 and p40 are expected to give a band of 2000bp if RFP was inserted and 1320 if nothing was inserted
 "
"