Team:Cambridge/Bioluminescence/Background
From 2010.igem.org
| Line 8: | Line 8: | ||
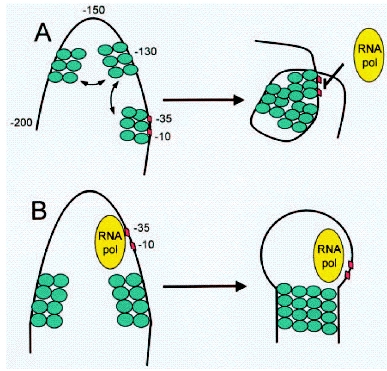
[[Image:H-NS.jpg|300px|thumb|left|H-NS repression via (A) promoter occlusion and (B) Polymerase trapping]] | [[Image:H-NS.jpg|300px|thumb|left|H-NS repression via (A) promoter occlusion and (B) Polymerase trapping]] | ||
A nucleoid protein, H-NS, appears to be intricately involved in the regulation of the transcription of Lux genes. H-NS is a pleiotropic repressor protein that has been shown to bind predominantly to curved DNA caused by A-T rich regions. Repression can occur both by binding to promoter sites (promoter occlusion) and by looping the DNA around a promoter region to trap the RNA-Polymerase ([http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=eurekah&part=A31864 Pon et al. 2000]) The H-NS protein consist of a dimerization (or multimerization) and a DNA-binding domain. Its binding to DNA appears to be DNA shape- rather than sequence-specific. A description of the DNA binding properties of H-NS proteins can be found in ([http://www.nature.com/nature/journal/v444/n7117/full/nature05283.html Dame et al. 2006]) The protein has also been implicated in the repression of foreign genes acquired by horizontal transfer and synthetic genes ([http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6976.2008.00155.x/pdf Dorman and Kane 2008]). Certain h-ns mutants have been shown to exhibit much higher levels of gene expression and bioluminescence. Several sites within the Lux system have been described as binding sites for H-NS ([http://onlinelibrary.wiley.com/doi/10.1002/(SICI)1099-1271(199807/08)13:4%3C185::AID-BIO486%3E3.0.CO;2-U/abstract Ulitzur 1998]). Such binding sites exist in both the leftward and the rightward promoter regions, within the coding sequence of LuxI, the intergenic region and start of LuxC as well as the intergenic region and start of LuxA. | A nucleoid protein, H-NS, appears to be intricately involved in the regulation of the transcription of Lux genes. H-NS is a pleiotropic repressor protein that has been shown to bind predominantly to curved DNA caused by A-T rich regions. Repression can occur both by binding to promoter sites (promoter occlusion) and by looping the DNA around a promoter region to trap the RNA-Polymerase ([http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=eurekah&part=A31864 Pon et al. 2000]) The H-NS protein consist of a dimerization (or multimerization) and a DNA-binding domain. Its binding to DNA appears to be DNA shape- rather than sequence-specific. A description of the DNA binding properties of H-NS proteins can be found in ([http://www.nature.com/nature/journal/v444/n7117/full/nature05283.html Dame et al. 2006]) The protein has also been implicated in the repression of foreign genes acquired by horizontal transfer and synthetic genes ([http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6976.2008.00155.x/pdf Dorman and Kane 2008]). Certain h-ns mutants have been shown to exhibit much higher levels of gene expression and bioluminescence. Several sites within the Lux system have been described as binding sites for H-NS ([http://onlinelibrary.wiley.com/doi/10.1002/(SICI)1099-1271(199807/08)13:4%3C185::AID-BIO486%3E3.0.CO;2-U/abstract Ulitzur 1998]). Such binding sites exist in both the leftward and the rightward promoter regions, within the coding sequence of LuxI, the intergenic region and start of LuxC as well as the intergenic region and start of LuxA. | ||
| + | |||
| + | |||
| + | <html> | ||
| + | <div style="text-align:center"> | ||
| + | <object width="640" height="480"> | ||
| + | <param name="movie" value="http://cambridgeigem.org/peter.swf"> | ||
| + | <embed src="http://cambridgeigem.org/peter.swf" width="640" height="480"> | ||
| + | </embed> | ||
| + | </object> | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
==Regulation by LuxR and LuxI== | ==Regulation by LuxR and LuxI== | ||
Revision as of 01:29, 27 October 2010

The Lux System
The Lux operon is a set of genes active in bacterial luminescence. Homologues are found in different species of luminescent bacteria, such as Vibrio fischeri, Vibrio harveyi, Vibrio (formerly Photobacterium) phosphoreum, Photobacterium leiognathi and Photorhabdus (Xenorhabdus) luminescens. Between these species there are slight differences in the order of genes. In the most studied species, V. fischeri, the system consists of two translated regions, a leftward region containing the LuxR gene and a rightward region containing the genes LuxI, C, D, A, B, E and G in this order. LuxA and LuxB encode the two subunits of the bacterial luciferase, while the products of LuxC, LuxD and LuxE synthesize the substrate for the light emitting reaction, tetradecanal. The exact function of LuxG is unknown, and it appears to be non-essential for light emission, but its presence increases light output. Due to the specific codon usage in the Lux operon, LuxA and LuxB are translated at a five times higher level than C, D, E and G. In V.fischeri and many other species, the expression of the Lux system is tightly coupled with the intra-specific quorum sensing mechanisms. Thus bioluminescence is controlled by the physiological state of the cell (availability of substrates) as well as the state of the colony (concentration of quorum-sensing molecules)
Repression by H-NS
A nucleoid protein, H-NS, appears to be intricately involved in the regulation of the transcription of Lux genes. H-NS is a pleiotropic repressor protein that has been shown to bind predominantly to curved DNA caused by A-T rich regions. Repression can occur both by binding to promoter sites (promoter occlusion) and by looping the DNA around a promoter region to trap the RNA-Polymerase ([http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=eurekah&part=A31864 Pon et al. 2000]) The H-NS protein consist of a dimerization (or multimerization) and a DNA-binding domain. Its binding to DNA appears to be DNA shape- rather than sequence-specific. A description of the DNA binding properties of H-NS proteins can be found in ([http://www.nature.com/nature/journal/v444/n7117/full/nature05283.html Dame et al. 2006]) The protein has also been implicated in the repression of foreign genes acquired by horizontal transfer and synthetic genes ([http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6976.2008.00155.x/pdf Dorman and Kane 2008]). Certain h-ns mutants have been shown to exhibit much higher levels of gene expression and bioluminescence. Several sites within the Lux system have been described as binding sites for H-NS ([http://onlinelibrary.wiley.com/doi/10.1002/(SICI)1099-1271(199807/08)13:4%3C185::AID-BIO486%3E3.0.CO;2-U/abstract Ulitzur 1998]). Such binding sites exist in both the leftward and the rightward promoter regions, within the coding sequence of LuxI, the intergenic region and start of LuxC as well as the intergenic region and start of LuxA.
Regulation by LuxR and LuxI
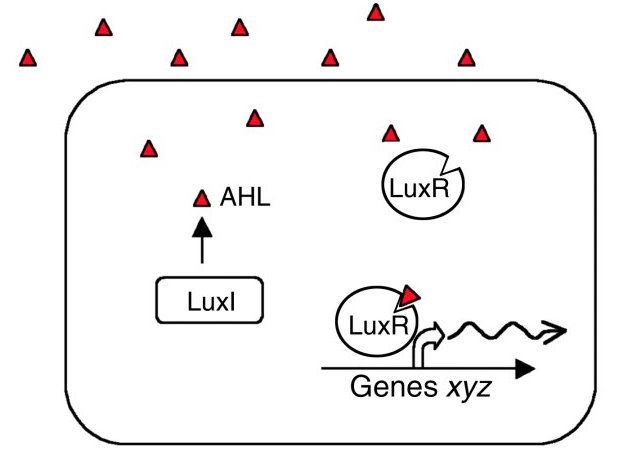
LuxR and LuxI are genes involved in the quorum sensing mechanism of Vibrio fischeri. LuxI codes for an enzyme catalysing the synthesis of a specific N-acyl homoserine lactone (AHL). This compound is diffusible and acts as a signal between different cells of the population. In nature, V. fischeri uses quorum sensing to assess the size of the symbiont colony within their host organism. At the right colony density they activate bioluminescence. AHL binds to the protein product of LuxR changing its 3-dimensional shape ([http://jb.asm.org/cgi/content/short/179/2/557 Stevens and Greenberg 1997]). The C-terminal domain of the activated LuxR then interacts with H-NS proteins bound to curved DNA-regions in the Lux operon. Such regions are especially prominent in the promoter regions of LuxI and LuxR, causing AHL to induce its own synthesis. Repression also occurs at the LuxCDABEG promoter as well as within the coding sequence of LuxC, LuxA and LuxB. While the natural quorum-controlled mechanism relieves this repression, it remains a problem if the lux operon is placed under a different promoter in a H-NS wild type strain.
 "
"