Team:ETHZ Basel/Achievements
From 2010.igem.org
(→MATLAB Toolbox (Lemming Toolbox)) |
|||
| Line 17: | Line 17: | ||
As a result of our efforts to make all our computer based tools & algorithms modular and re-usable, we have assembled the entire ''in silico'' setup of the E. Lemming into a [https://2010.igem.org/Team:ETHZ_Basel/Achievements/Matlab_Toolbox MATLAB Toolbox] that includes 18 Simulink blocks that encapsulate the algorithms we developed/implemented. This is available for [http://sourceforge.net/projects/ethzigem10/files/LemmingToolbox_Setup.zip/download downloading]. | As a result of our efforts to make all our computer based tools & algorithms modular and re-usable, we have assembled the entire ''in silico'' setup of the E. Lemming into a [https://2010.igem.org/Team:ETHZ_Basel/Achievements/Matlab_Toolbox MATLAB Toolbox] that includes 18 Simulink blocks that encapsulate the algorithms we developed/implemented. This is available for [http://sourceforge.net/projects/ethzigem10/files/LemmingToolbox_Setup.zip/download downloading]. | ||
| + | |||
| + | == Technical Standard == | ||
== Model of light-inducible chemotaxis pathway == | == Model of light-inducible chemotaxis pathway == | ||
| Line 23: | Line 25: | ||
== Movement Model == | == Movement Model == | ||
We made a [https://2010.igem.org/Team:ETHZ_Basel/Modeling/Movement stochastic model to realistically simulate the chemotaxis motion] of the cell, which responds to the light input that is converted into changes in the chemotaxis pathway, that influence the motion of the cell. | We made a [https://2010.igem.org/Team:ETHZ_Basel/Modeling/Movement stochastic model to realistically simulate the chemotaxis motion] of the cell, which responds to the light input that is converted into changes in the chemotaxis pathway, that influence the motion of the cell. | ||
| + | |||
| + | == Microscope Control == | ||
| + | |||
| + | == Lemming Game == | ||
== Systems Design == | == Systems Design == | ||
== Systems Implementation == | == Systems Implementation == | ||
| - | |||
| - | |||
Revision as of 15:40, 26 October 2010
Achievements Overview
BioBrick Toolbox
Being now BBF RFC28 (cloning strategy for combinatorial multip-part assembly [http://dspace.mit.edu/handle/1721.1/46721] ) experts, we highly recommend you to check out our BrickBox collection comprising of proteins used for chemotaxis, cellular anchoring, light sensing and fluorescent reporting. We also gave a lot of thoughts how to make BBF RFC28 (method for combinatorial multi-part assembly based on the Type II restriction enzyme AarI) compatible with Tom Knight's OAS standard.
Also about the characterization of our parts you will find more information on this page.
Don't forget our favourite parts including the light sensing couple PhyB-Pif3 and the Archaeal light receptor!
MATLAB Toolbox (Lemming Toolbox)
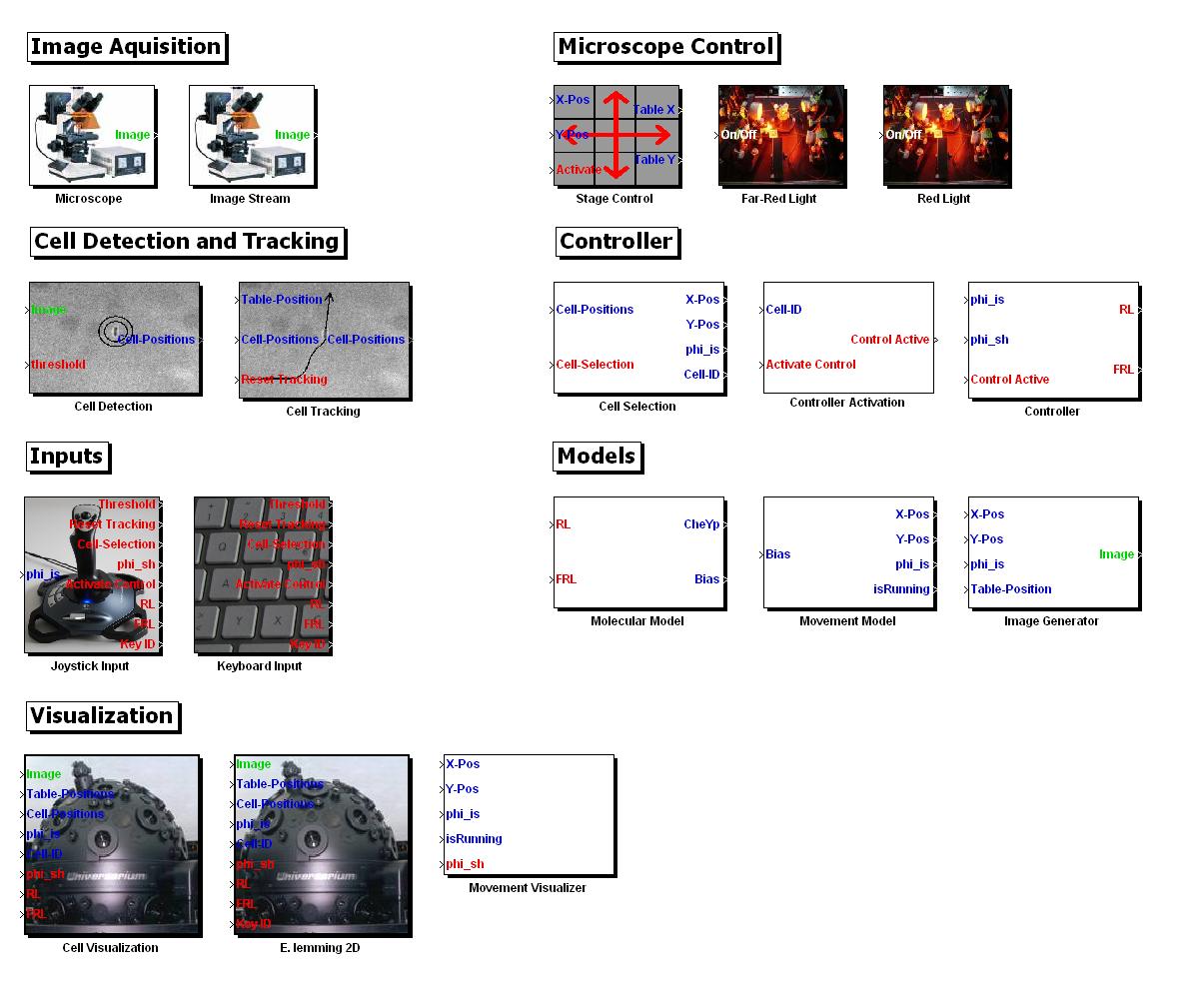
As a result of our efforts to make all our computer based tools & algorithms modular and re-usable, we have assembled the entire in silico setup of the E. Lemming into a MATLAB Toolbox that includes 18 Simulink blocks that encapsulate the algorithms we developed/implemented. This is available for [http://sourceforge.net/projects/ethzigem10/files/LemmingToolbox_Setup.zip/download downloading].
Technical Standard
Model of light-inducible chemotaxis pathway
We made a model to control the chemotaxis by uisng light. The idea was to use Light Seceptive Proteins to change the concentrations of the proteins responsible for chemotaxis, in such a way that the motion of the cell can be controlled the way we want.
Movement Model
We made a stochastic model to realistically simulate the chemotaxis motion of the cell, which responds to the light input that is converted into changes in the chemotaxis pathway, that influence the motion of the cell.
 "
"


