Team:UNIPV-Pavia/Project/results
From 2010.igem.org
(→Quantification of the HSL produced) |
(→Results) |
||
| Line 309: | Line 309: | ||
|- | |- | ||
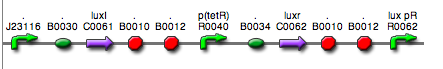
|<partinfo>BBa_K300030</partinfo> in <partinfo>pSB1A2</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23118 | |<partinfo>BBa_K300030</partinfo> in <partinfo>pSB1A2</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23118 | ||
| - | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A2</partinfo><br>[[Image:pv_T9002.png|300px]] | + | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A2</partinfo><br>[[Image:pv_T9002.png|300px]] |
|<partinfo>BBa_K300024</partinfo><br>in <partinfo>pSB1A2</partinfo> | |<partinfo>BBa_K300024</partinfo><br>in <partinfo>pSB1A2</partinfo> | ||
|- | |- | ||
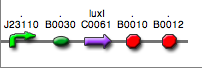
|<partinfo>BBa_K300028</partinfo> in <partinfo>pSB1A2</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23110 | |<partinfo>BBa_K300028</partinfo> in <partinfo>pSB1A2</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23110 | ||
| - | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A2</partinfo><br>[[Image:pv_T9002.png|300px]] | + | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A2</partinfo><br>[[Image:pv_T9002.png|300px]] |
|<partinfo>BBa_K300021</partinfo><br>in <partinfo>pSB1A2</partinfo> | |<partinfo>BBa_K300021</partinfo><br>in <partinfo>pSB1A2</partinfo> | ||
|- | |- | ||
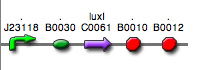
|<partinfo>BBa_K300029</partinfo> in <partinfo>pSB1A2</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23116 | |<partinfo>BBa_K300029</partinfo> in <partinfo>pSB1A2</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23116 | ||
| - | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A2</partinfo><br>[[Image:pv_T9002.png|300px]] | + | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A2</partinfo><br>[[Image:pv_T9002.png|300px]] |
|<partinfo>BBa_K300022</partinfo><br>in <partinfo>pSB1A2</partinfo> | |<partinfo>BBa_K300022</partinfo><br>in <partinfo>pSB1A2</partinfo> | ||
|- | |- | ||
|<partinfo>BBa_K300026</partinfo> in <partinfo>pSB1A2</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23105 | |<partinfo>BBa_K300026</partinfo> in <partinfo>pSB1A2</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23105 | ||
| - | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A2</partinfo><br>[[Image:pv_T9002.png|300px]] | + | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A2</partinfo><br>[[Image:pv_T9002.png|300px]] |
|<partinfo>BBa_K300019</partinfo><br>in <partinfo>pSB1A2</partinfo> | |<partinfo>BBa_K300019</partinfo><br>in <partinfo>pSB1A2</partinfo> | ||
|- | |- | ||
|xxx<br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23114 | |xxx<br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23114 | ||
| - | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A2</partinfo><br>[[Image:pv_T9002.png|300px]] | + | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A2</partinfo><br>[[Image:pv_T9002.png|300px]] |
|<partinfo>BBa_K300023</partinfo><br>in <partinfo>pSB1A2</partinfo> | |<partinfo>BBa_K300023</partinfo><br>in <partinfo>pSB1A2</partinfo> | ||
|- | |- | ||
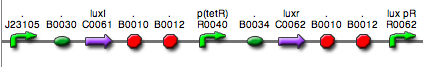
|<partinfo>BBa_K300030</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23118 | |<partinfo>BBa_K300030</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23118 | ||
| - | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_T9002.png|300px]] | + | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_T9002.png|300px]] |
|<partinfo>BBa_K300024</partinfo><br>in <partinfo>pSB4C5</partinfo> | |<partinfo>BBa_K300024</partinfo><br>in <partinfo>pSB4C5</partinfo> | ||
|- | |- | ||
|<partinfo>BBa_K300028</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23110 | |<partinfo>BBa_K300028</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23110 | ||
| - | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_T9002.png|300px]] | + | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_T9002.png|300px]] |
|<partinfo>BBa_K300021</partinfo><br>in <partinfo>pSB4C5</partinfo> | |<partinfo>BBa_K300021</partinfo><br>in <partinfo>pSB4C5</partinfo> | ||
|- | |- | ||
|<partinfo>BBa_K300029</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23116 | |<partinfo>BBa_K300029</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23116 | ||
| - | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_T9002.png|300px]] | + | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_T9002.png|300px]] |
|<partinfo>BBa_K300022</partinfo><br>in <partinfo>pSB4C5</partinfo> | |<partinfo>BBa_K300022</partinfo><br>in <partinfo>pSB4C5</partinfo> | ||
|- | |- | ||
|<partinfo>BBa_K300026</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23105 | |<partinfo>BBa_K300026</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23105 | ||
| - | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_T9002.png|300px]] | + | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_T9002.png|300px]] |
|<partinfo>BBa_K300019</partinfo><br>in <partinfo>pSB4C5</partinfo> | |<partinfo>BBa_K300019</partinfo><br>in <partinfo>pSB4C5</partinfo> | ||
|- | |- | ||
|<partinfo>BBa_K300030</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23118 | |<partinfo>BBa_K300030</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23118 | ||
| - | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A3</partinfo><br>[[Image:pv_T9002.png|300px]] | + | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A3</partinfo><br>[[Image:pv_T9002.png|300px]] |
|Sender and Receiver are contained in two different plasmids, <br>cotransformed in the same cell | |Sender and Receiver are contained in two different plasmids, <br>cotransformed in the same cell | ||
|- | |- | ||
|<partinfo>BBa_K300028</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23110 | |<partinfo>BBa_K300028</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23110 | ||
| - | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A3</partinfo><br>[[Image:pv_T9002.png|300px]] | + | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A3</partinfo><br>[[Image:pv_T9002.png|300px]] |
|Sender and Receiver are contained in two different plasmids, <br>cotransformed in the same cell | |Sender and Receiver are contained in two different plasmids, <br>cotransformed in the same cell | ||
|- | |- | ||
|<partinfo>BBa_K300029</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23116 | |<partinfo>BBa_K300029</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23116 | ||
| - | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A3</partinfo><br>[[Image:pv_T9002.png|300px]] | + | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A3</partinfo><br>[[Image:pv_T9002.png|300px]] |
|Sender and Receiver are contained<br> in two different plasmids, <br>cotransformed in the same cell | |Sender and Receiver are contained<br> in two different plasmids, <br>cotransformed in the same cell | ||
|- | |- | ||
|<partinfo>BBa_K300026</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23105 | |<partinfo>BBa_K300026</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23105 | ||
| - | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A3</partinfo><br>[[Image:pv_T9002.png|300px]] | + | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A3</partinfo><br>[[Image:pv_T9002.png|300px]] |
|Sender and Receiver are contained<br> in two different plasmids, <br>cotransformed in the same cell | |Sender and Receiver are contained<br> in two different plasmids, <br>cotransformed in the same cell | ||
|- | |- | ||
|<partinfo>BBa_K300025</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23101 | |<partinfo>BBa_K300025</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23101 | ||
| - | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A3</partinfo><br>[[Image:pv_T9002.png|300px]] | + | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A3</partinfo><br>[[Image:pv_T9002.png|300px]] |
|Sender and Receiver are contained<br> in two different plasmids, <br>cotransformed in the same cell | |Sender and Receiver are contained<br> in two different plasmids, <br>cotransformed in the same cell | ||
|- | |- | ||
|<partinfo>BBa_K300027</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23106 | |<partinfo>BBa_K300027</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23106 | ||
| - | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A3</partinfo><br>[[Image:pv_T9002.png|300px]] | + | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A3</partinfo><br>[[Image:pv_T9002.png|300px]] |
|Sender and Receiver are contained<br> in two different plasmids, <br>cotransformed in the same cell | |Sender and Receiver are contained<br> in two different plasmids, <br>cotransformed in the same cell | ||
|} | |} | ||
Revision as of 08:57, 26 October 2010
|
Self-inducible promotersRegulation of signal protein productionExperimental implementation: <partinfo>BBa_K300009</partinfo> part was assembled downstream of different constitutive promoters, thus obtaining a signal molecule generator. The choice of constitutive promoters was performed between the ones belonging to the [http://partsregistry.org/Part:BBa_J23101 Anderson’s promoters collection] ; we chose promoters according to their activities reported in the Registry of Standard Biological Parts, in order to have a thick mesh:
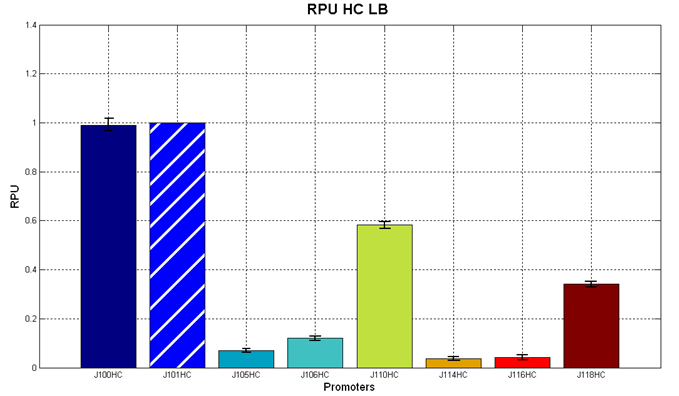
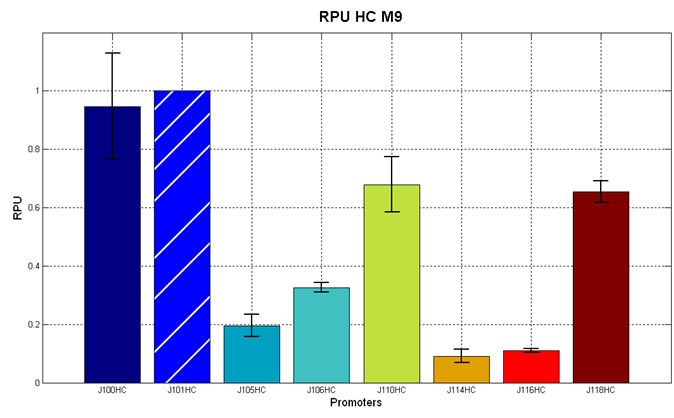
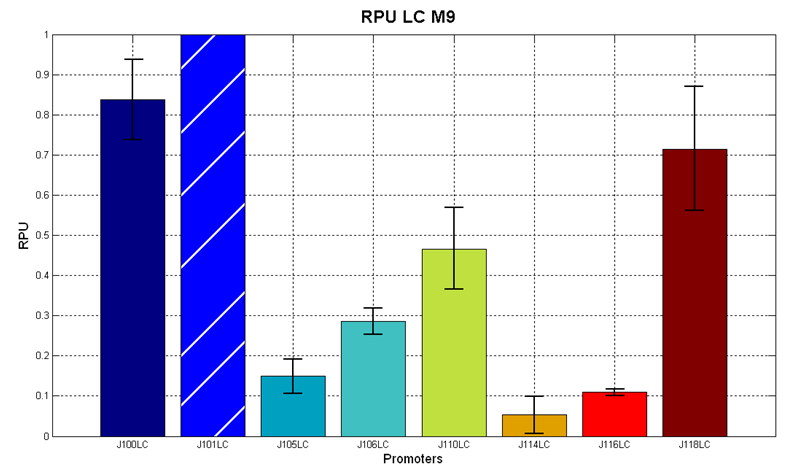
Before constructing the signal generators, <partinfo>BBa_K300009</partinfo> and <partinfo>BBa_K300010</partinfo> under the regulation of one of these constitutive promoters, we evaluated the promoter activities in Relative Promoter Units (R.P.U.) according to Data analysis for RPU evaluation, using the reporter protein RFP (Red Fluorescent Protein) in different experimental conditions (plasmids’ copy number and growth medium), many of them not yet explored and documented:
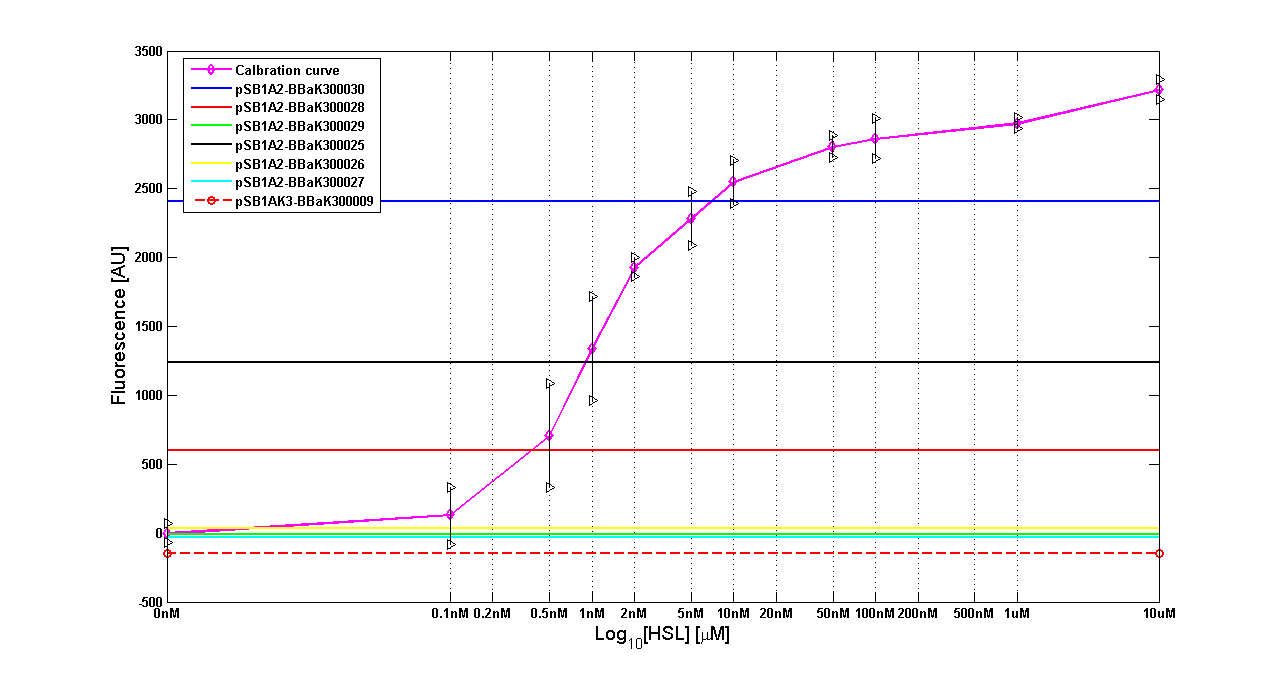
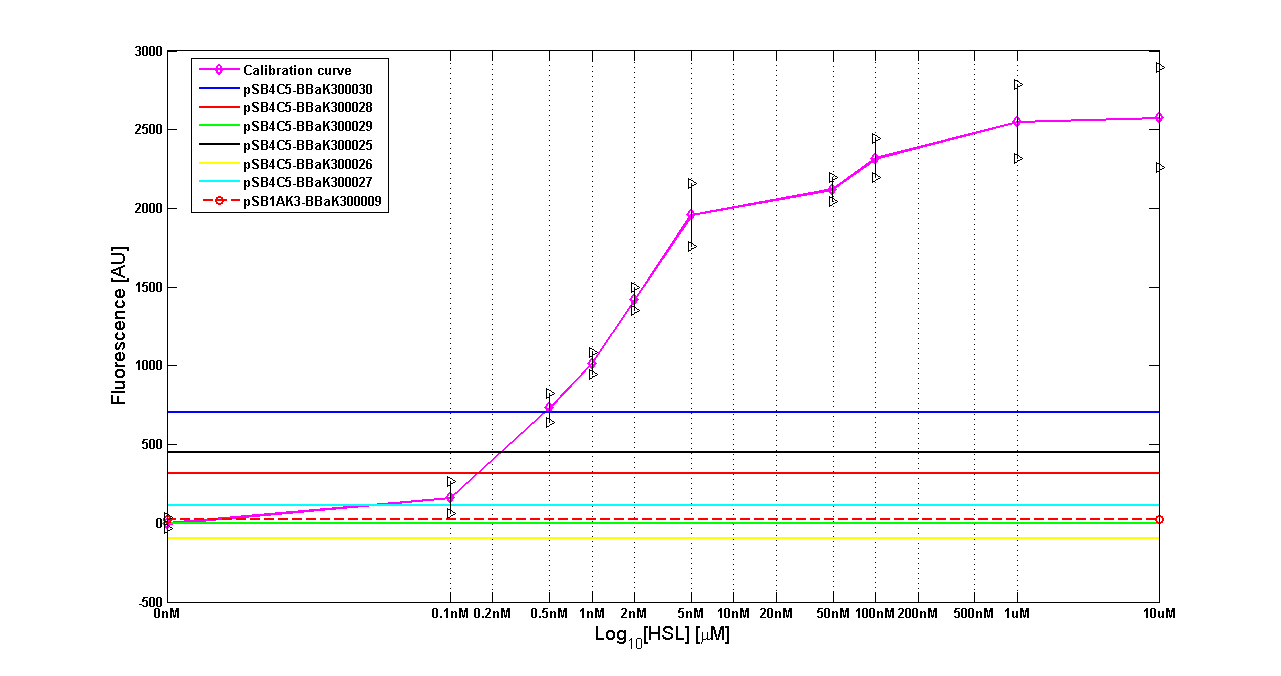
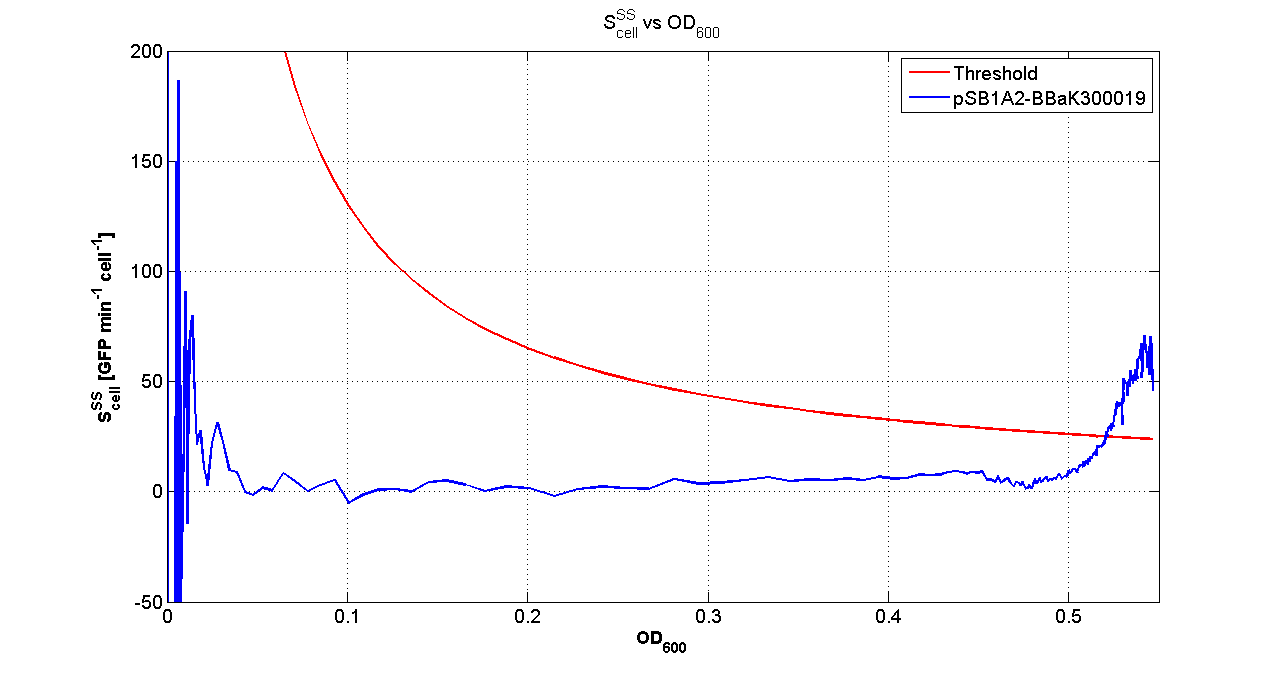
It was not possible to evaluate promoters activities in low copy number plasmids and LB because the RFP activity was too weak and not distinguishable from the background. RFP fluorescence and Optical Density at 600nm (O.D.600) were measured in 96-well microplates, as reported in Microplate reader experiments for constitutive promoters (R.P.U. evaluation) - Protocol #2 and data were analyzed as reported in Data Analysis RPU; Results: results are shown here. Discussion: we observed that the ranking previously documented in the Registry is not valid in all the conditions, even if a general agreement can be observed. As an example, <partinfo>BBa_J23110</partinfo> in high copy plasmid is stronger than <partinfo>BBa_J23118</partinfo>, in contrast with the ranking reported in the Registry. After the evaluation of promoter activity, signal generators were constructed in high copy and low copy plasmids: <partinfo>BBa_K300009</partinfo> and <partinfo>BBa_K300010</partinfo> were assembled downstream of the above mentioned promoters, thus obtaining the following parts: Some of the promoters could not be cloned upstream of these devices because they produced LuxI protein amounts that give a high metabolic burden for E. coli, so it was not possible to study all the combinations as transformans could not be obtained in some cases. For each part, a measurement system was built, exploiting the production of the reporter gene GFP (Green Fluoresent Protein) to evaluate the "switch on" condition of every self-inducible promoter. Many different combinations were explored, in order to provide a library of promoters able to initiate transcription at the desired culture density. Quantification of the HSL producedExperimental implementation The new parts were, thus, characterized, measuring the HSL concentration released in the medium after a 6 hour growth of the cultures. All the details are available in this section. <partinfo>BBa_T9002</partinfo> contained in <partinfo>pSB1A3</partinfo> in E. coli TOP10 was used as a HSL->GFP biosensor. In every experiment, a HSL-GFP calibration curve with known concentration of HSL was produced. Results The amount of 3OC6-HSL produced after a 6 hours growth by E. coli DH5alpha bearing the parts contained in high copy plasmid <partinfo>pSB1A2</partinfo> is reported in Fig.8 and in the table:
The amount of 3OC6-HSL produced after 6 hours growth by the parts contained in low copy plasmid <partinfo>pSB4C5</partinfo> is reported in Fig.9 and in the table:
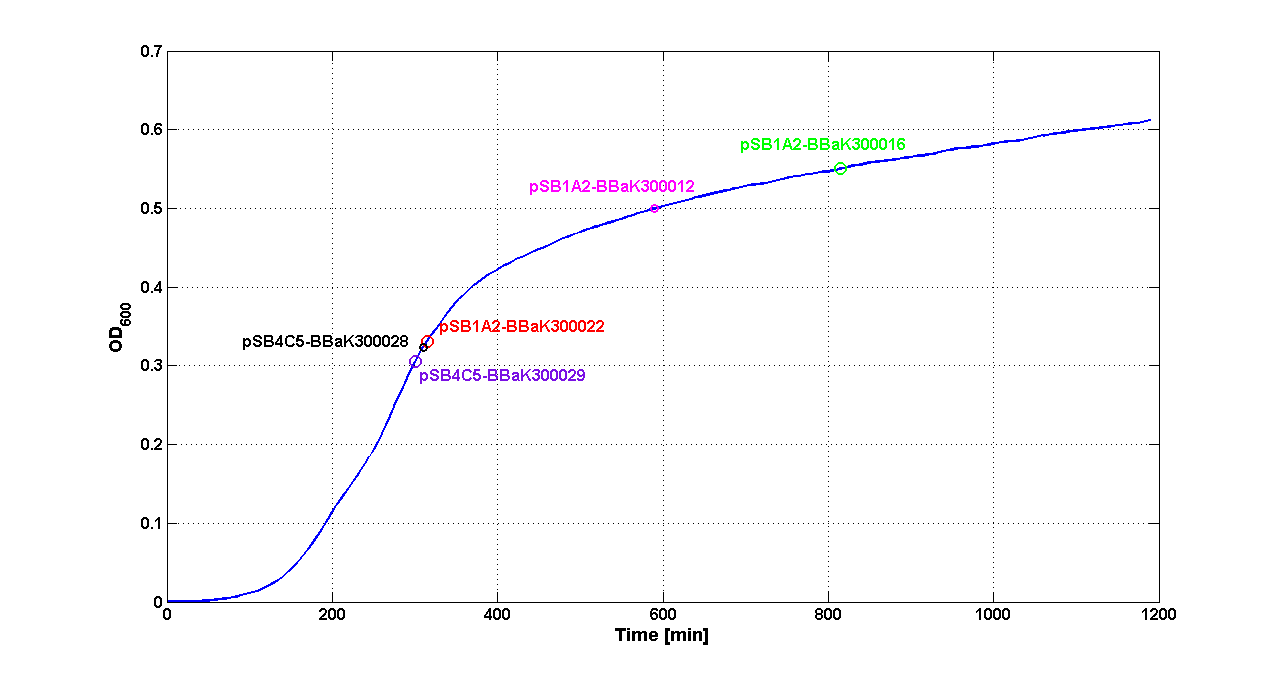
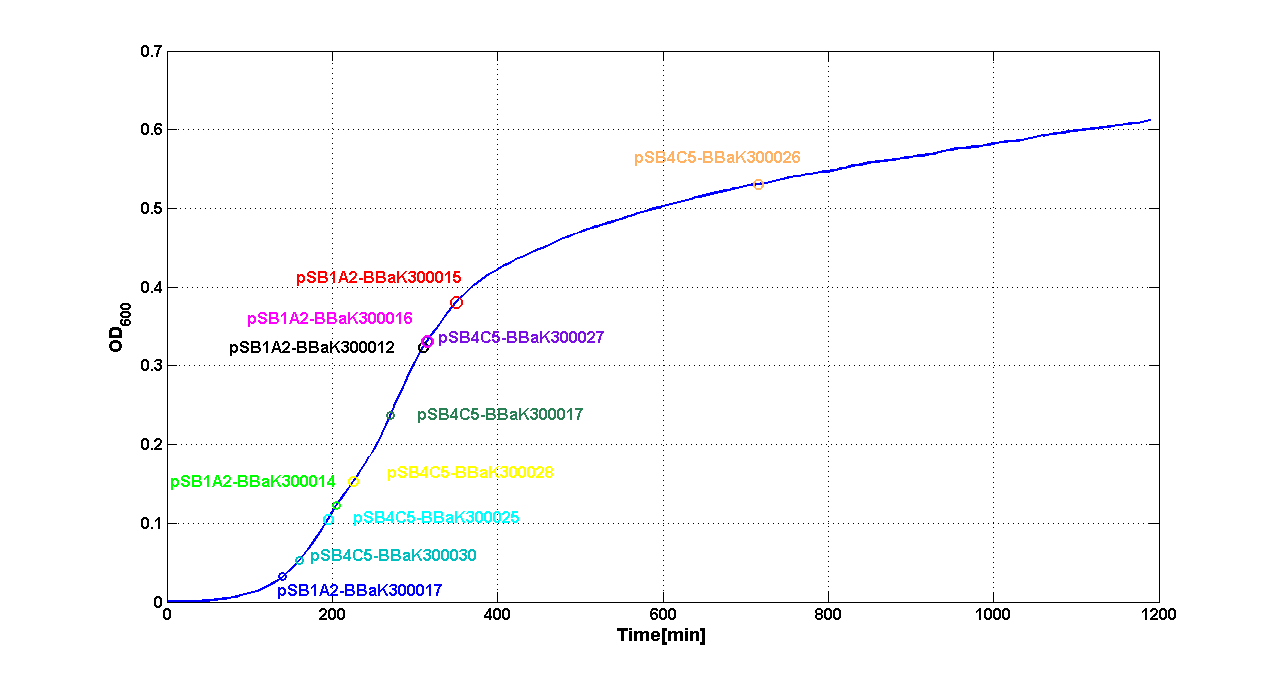
Discussion These experiments provided extremely useful informations about the capability of the signal generators to produce the 3OC6-HSL signal molecule. Data are quantitative, but incomplete because for weak promoters or medium-strength promoters contained in a low copy number plasmid the amount of 3OC6-HSL was not detectable using this system. However, this simple experiment shows that there is a strong correlation between the strength of promoter and the amount of signal molecule produced. These results confirm that the production of the autoinducer can be engineered in E. coli and different expression systems reach different amounts of 3OC6-HSL in the growth media as a function of the promoter strength. Thus, these results demonstrate that self-inducible circuits can be rationally designed from a set of well characterized standard parts. Modulation of plasmid copy numberSignal generator and sensor device were assembled in an unique part (such as <partinfo>BBa_K300017</partinfo>, <partinfo>BBa_K300014</partinfo>, <partinfo>BBa_K300015</partinfo>, <partinfo>BBa_K300016</partinfo> and <partinfo>BBa_K300012</partinfo>) beared on high copy number plasmid <partinfo>pSB1A2</partinfo> or low copy number plasmid <partinfo>pSb4C5</partinfo>. A third alternative was the assembly of signal generator on a low copy number plasmid (<partinfo>pSB4C5</partinfo>) and the receiver device on high number plasmid (<partinfo>pSB1A2</partinfo>). The circuits we obtained and tested are summarized here:
ResultsThe following measurement systems were realized assembling GFP downstream of each self-inducible device. The parts characterized are reported in this table: Cultures of E. coli TOP10 bearing the plasmids containing the self-inducible devices expressing G.F.P. were grown according to this protocol and all data collected were analyzed as explained in this section Doubling times were estimated as explained here Thus, these BioBrick parts can be used to express recombinant proteins without adding an inducer to trigger the transcription of their genes; in large-scale production of such proteins this strategy could be also cost saving. For every self-inducible device, several parameters were evaluated, as reported in this section. Results are summarized in the following tables:
Tab. 2 - Sender and Receiver on low copy plasmid <partinfo>pSB4C5</partinfo> Tab. 3 - Sender on low copy plasmid <partinfo>pSB4C5</partinfo> and Receiver on high copy plasmid <partinfo>pSB1A3</partinfo>
Integrative standard vector for E. coliMaterials and MethodsPlasmids and strains: the <partinfo>BBa_J72008</partinfo> helper plasmid was kindly given by Prof. JC Anderson (UC Berkeley). MC1061 (<partinfo>BBa_K300078</partinfo>) and MG1655 (<partinfo>BBa_V1000</partinfo>) E. coli strains and the pCP20 helper plasmid were purchased from the Coli Genetic Stock Center (Yale University). DH5alpha (<partinfo>BBa_V1001</partinfo>) strain was purchased from Invitrogen.
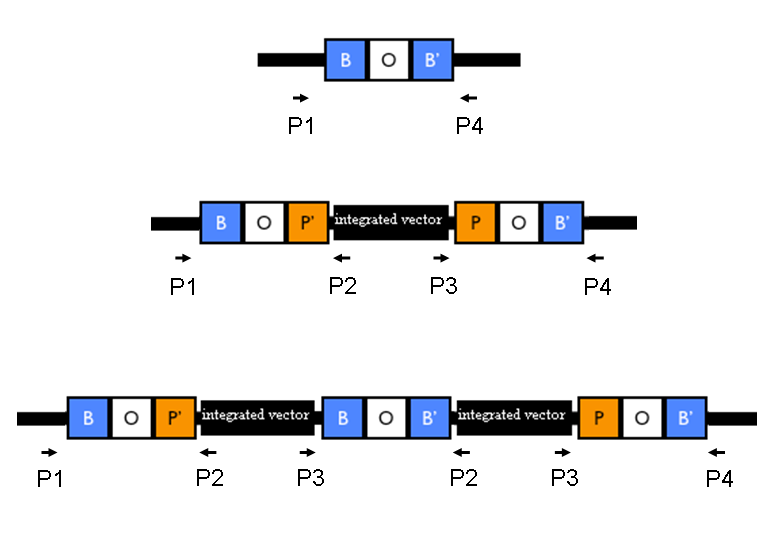
The relative position of the P1, P2, P3 and P4 primers is shown in Fig.1: Competent cells preparation: all the E. coli strains were made competent following a slightly modified version of the protocol described in [Sambrook J et al., 1989]. Briefly, cells were grown to and OD600 of ~0.4-0.6, harvested (4000 rpm, 10 min, 4°C) and the supernatant discarded. Cells were resuspended in (30 ml for each 50 ml of initial culture) pre-chilled Mg-Ca buffer (80 mM MgCl2, 20 mM CaCl2), centrifuged as before and the supernatant discarded. Cells were resuspended in (2 ml for each 50 ml of initial culture) pre-chilled Ca buffer (100 mM CaCl2, 15% glycerol), aliquoted in 0.5 ml tubes and freezed immediately at -80°C. Test the transformation efficiency as:
efficiency [CFU/ug of DNA]= # CFU * 1000 ng of DNA / amount of transformed DNA [ng]
Self-ligated <partinfo>BBa_K300008</partinfo> was prepared by digesting <partinfo>pSB1A2</partinfo>-<partinfo>BBa_K300008</partinfo> (yielded by BioBrick Standard Assembly) with XbaI-SpeI. The insert was isolated and purified from a 1% agarose gel. Then, it was self-ligated to generate a Cm-resistant R6K plasmid. The colonies were counted in each plate and the transformation efficiency was estimated as described before. The Chloramphenicol concentration in plates was 34 ug/ml for the high copy plasmids, 12.5 ug/ml for the medium/low copy plasmids and 12.5 for the three control strains transformed with the R6K plasmid.
All the DNA manipulations were performed according to manufacturer's protocols.
Validate the loss of the helper plasmid by inoculating colonies in Cm (at 12.5 ug/ml) media and counterselecting them in Amp (at 50 ug/ml) media. Validate the correct integration position by performing colony PCR with primers P1/P2, P3/P4, P1/P4, P2,P3 and VF2/VR. Validate the phenotype (when possible).
Validate the loss of the helper plasmid by inoculating colonies in Amp (at 100 ug/ml) media and validate the loss of the Cm resistance from the genome by inoculating colonies in Cm (at 12.5 ug/ml) media. Validate the correct length of the integrated part without Cm resistance and R6K origin by performing colony PCR with primers P1/P4 (which amplify the entire Phi80 locus) and VF2/VR (which amplify the integrated part). Validate the phenotype (when possible).
ResultsValidation of pir strains to propagate the R6K replication originThe BW25141 (pir+) and BW23474 (pir-116) E. coli strains were chosen to propagate the vectors with the R6K replication origin at medium copy (~15 molecules per cell) and high copy (~250) respectively. The results about their capability to propagate R6K plasmids are shown here:
For this reason, competent cells were prepared again for MG1655 and the transformation procedure was repeated for this strain, yielding no colonies in the <partinfo>BBa_K300008</partinfo> plate as expected. Miniprep of the BW25141 and BW23474 strains transformed with <partinfo>BBa_K300008</partinfo> yielded a DNA concentration of ~20 ng/ul (qualitatively comparable with medium copy number plasmids) and ~90-100 ng/ul (qualitatively comparable with high copy number plasmids). The results shown in the table above also show that the R6K plasmid in pir+ and pir-116 strains was transformed with the same efficiency as the pSB*** positive control plasmid, demonstrating that the R6K origin doesn't give any handicap in plasmid transformation. So, the BW25141 and BW23474 strains can be successfully used to propagate the integrative vector after the excision of the pUC19-derived high copy replication origin, present in the default insert <partinfo>BBa_I52002</partinfo>. Integration of the desired BioBrick part into the Phi80 genome locusMC1061 and MG1655 were chosen as host strains for integration. <partinfo>BBa_K173001</partinfo> (constitutive strong promoter with GFPmut3) and the EcoRI-PstI fragment of <partinfo>BBa_J61002</partinfo>-<partinfo>BBa_J23101</partinfo> (here called PconRFP - constitutive strong promoter with RFP) were chosen as two proof of concept BioBrick parts to test the integration capability of the <partinfo>BBa_K300000</partinfo> vector in the Phi80 genome locus of these strains. For this reason, <partinfo>BBa_K173001</partinfo> and PconRFP were ligated in <partinfo>BBa_K300000</partinfo> (digested with EcoRI-PstI) and propagated using BW23474. The integration protocol was performed as described in the Materials and Methods section for 4 different combination:
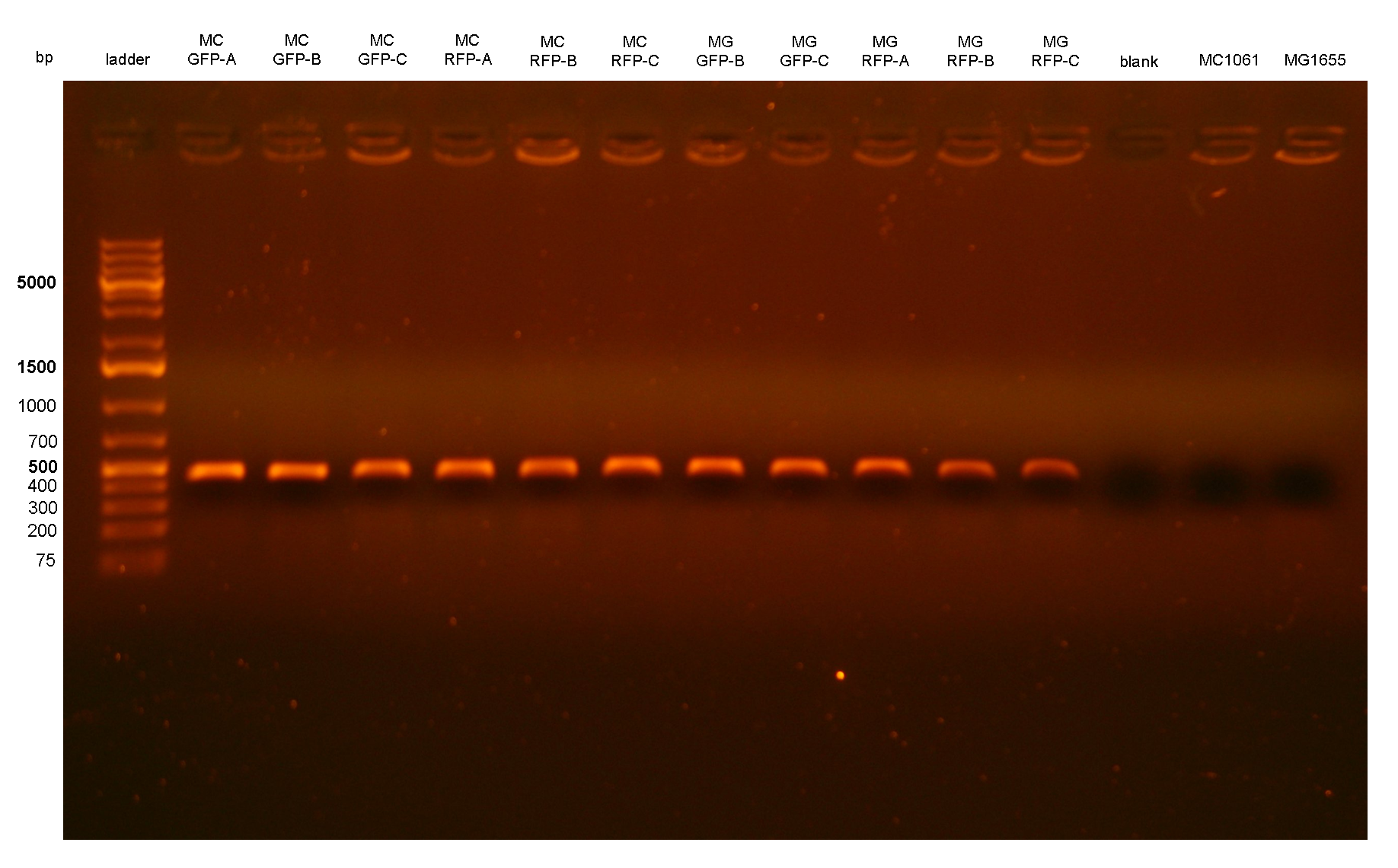
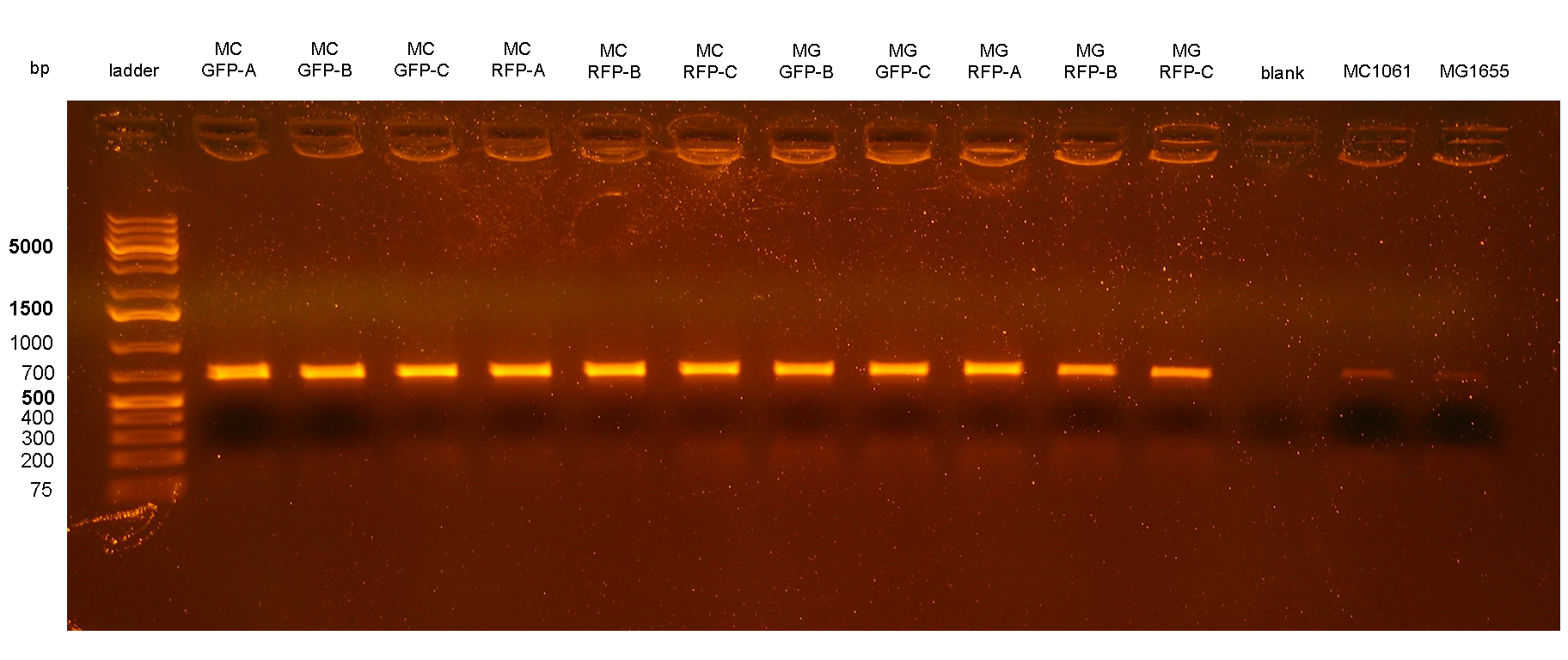
Three colonies grown after the overnight incubation at 43°C (step 5 of integration protocol) were analyzed for each plate. These 12 clones were called: MC-GFP-A,B,C , MC-RFP-A,B,C , MG-GFP-A,B,C and MG-RFP-A,B,C.
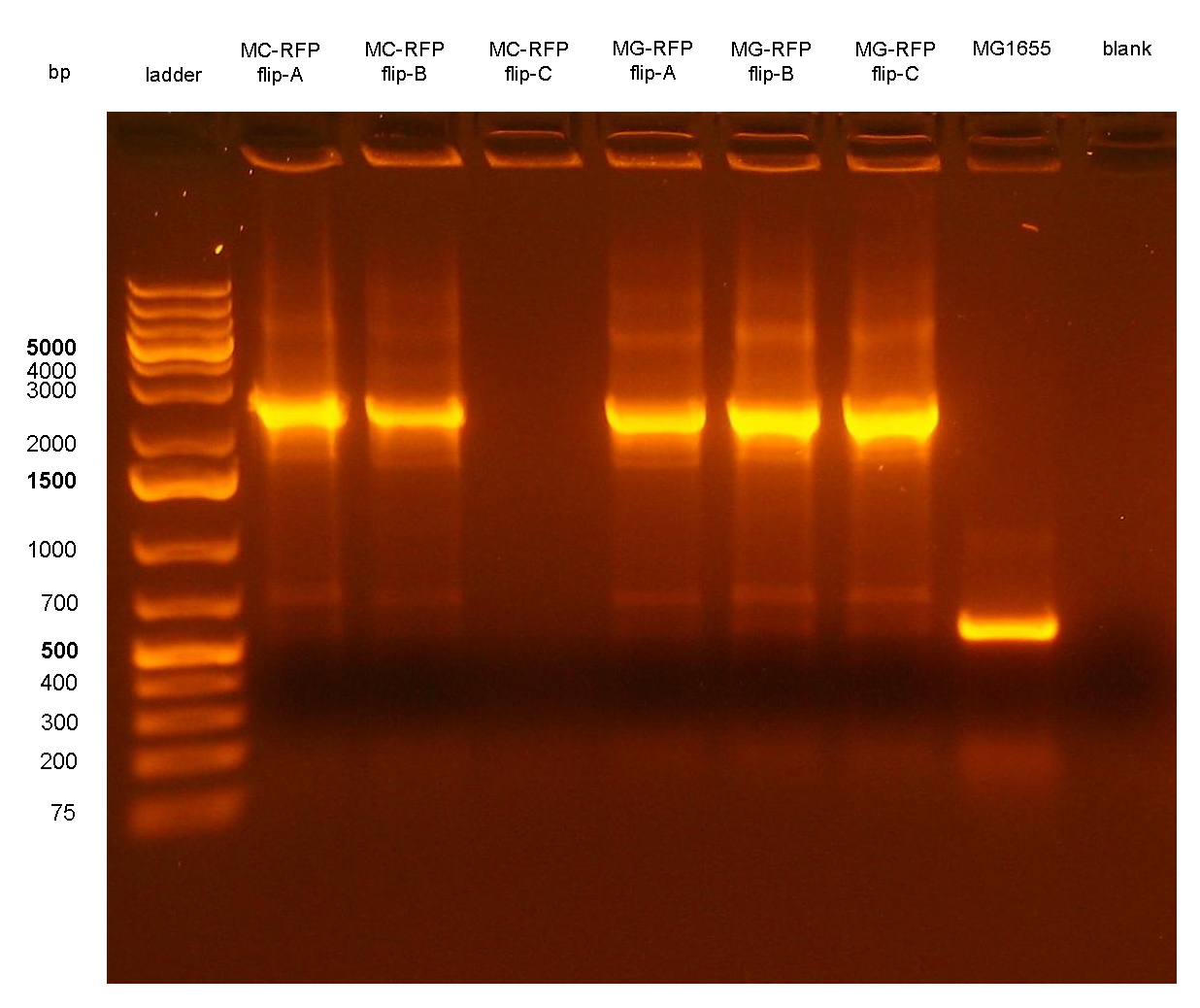
PCR results with primers P2/P3 showed that two clones (MC-GFP-B and MC-GFP-C) were single integrants, while all the other clones were multiple tandem integrants (i.e. the Phi80 locus contained more than one copy of the DNA of interest). Negative controls showed no amplicons, as expected.
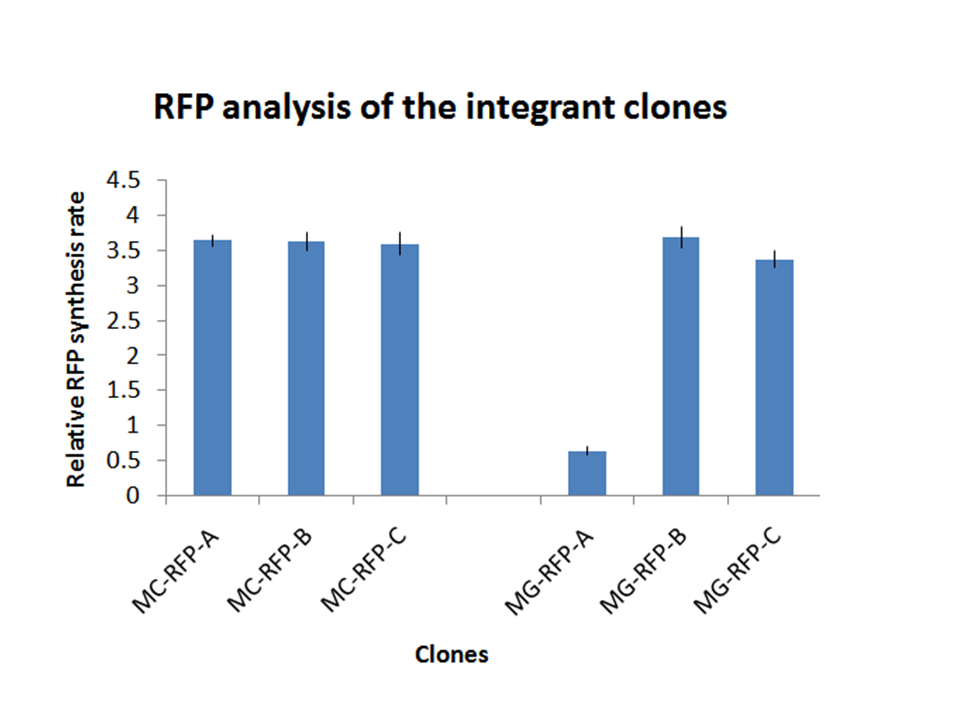
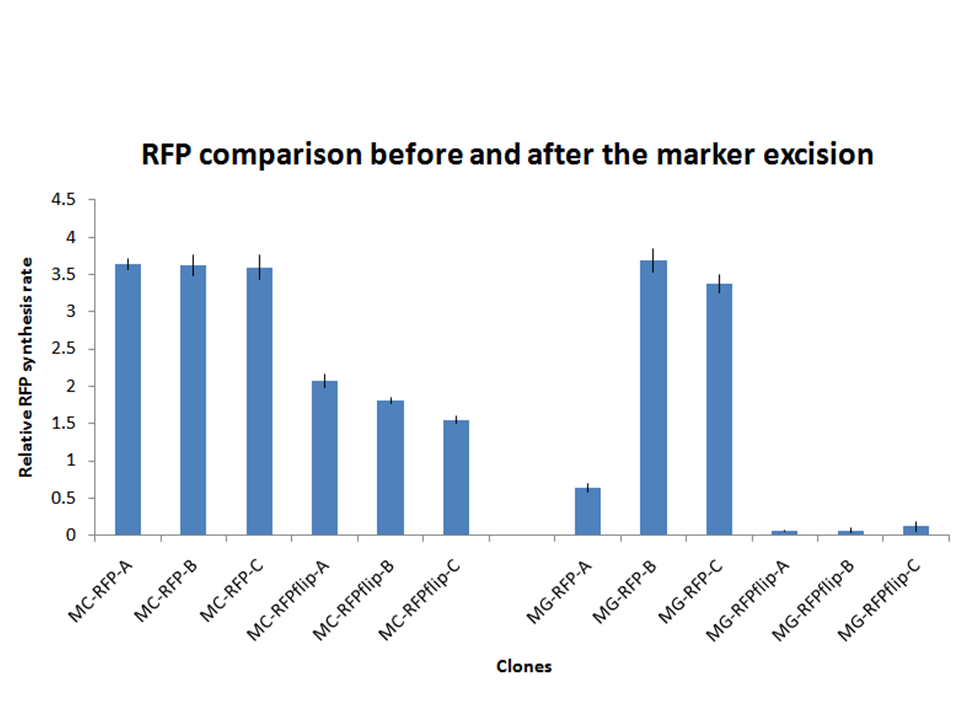
On the other hand, RFP clones (MC-RFP-A,B,C and MG-RFP-A,B,C) all showed a higher fluorescence than the negative controls (see Fig.5). As Fig.5 show, the fluorescence of the three MG-RFP had a higher variability between clones when compared to the three MC-RFP. However, the clones were not necessarily expected to behave in the same way because all of them were multiple tandem integrants and the copy number of the PconRFP construct could be arbitrary.
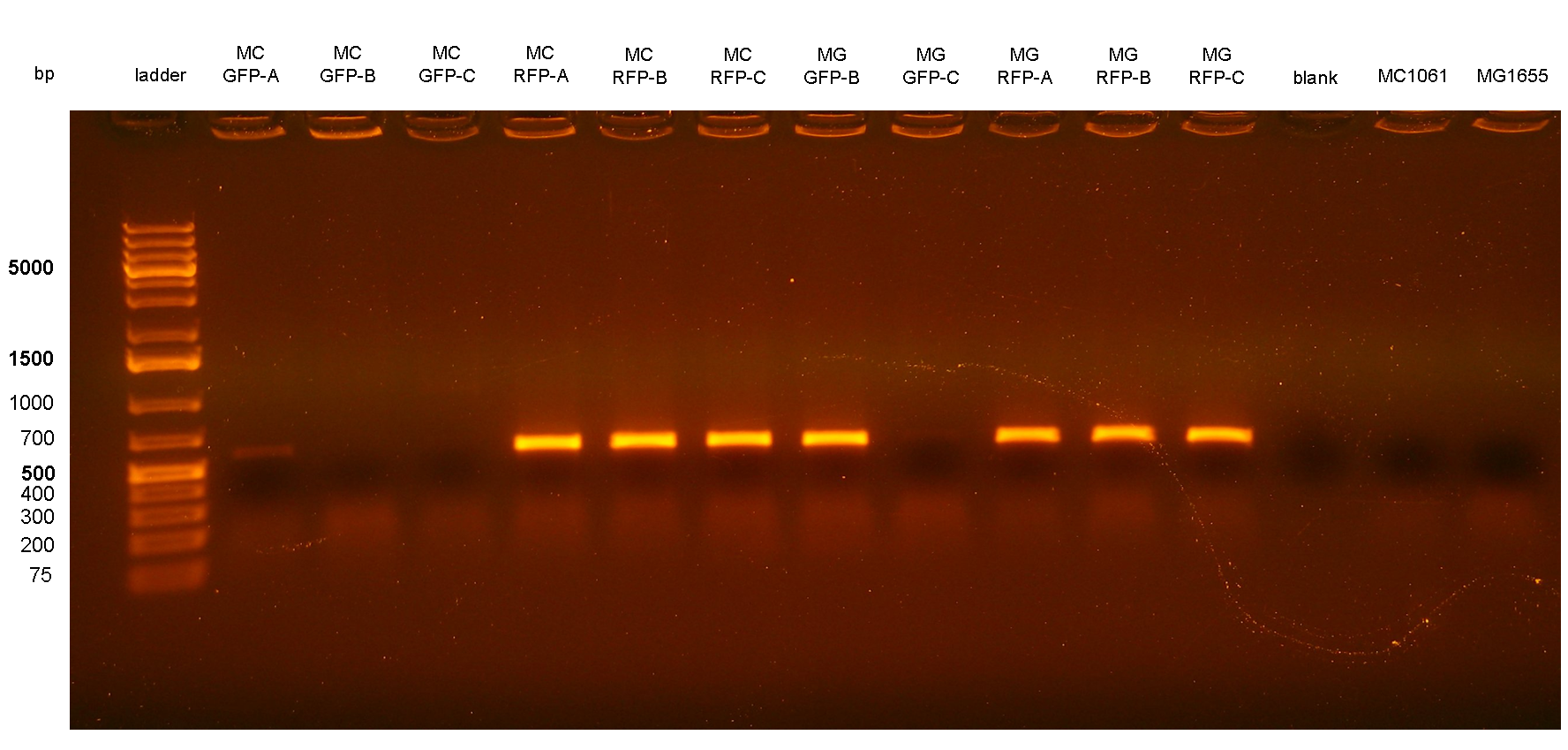
Chloramphenicol resistance marker excisionThe marker excision was performed on two of the previously validated integrant strains: MC-RFP-A and MG-RFP-A (even if they were multiple tandem integrants). The marker excision protocol was performed as described in the Materials and Methods section for both strains, here named:
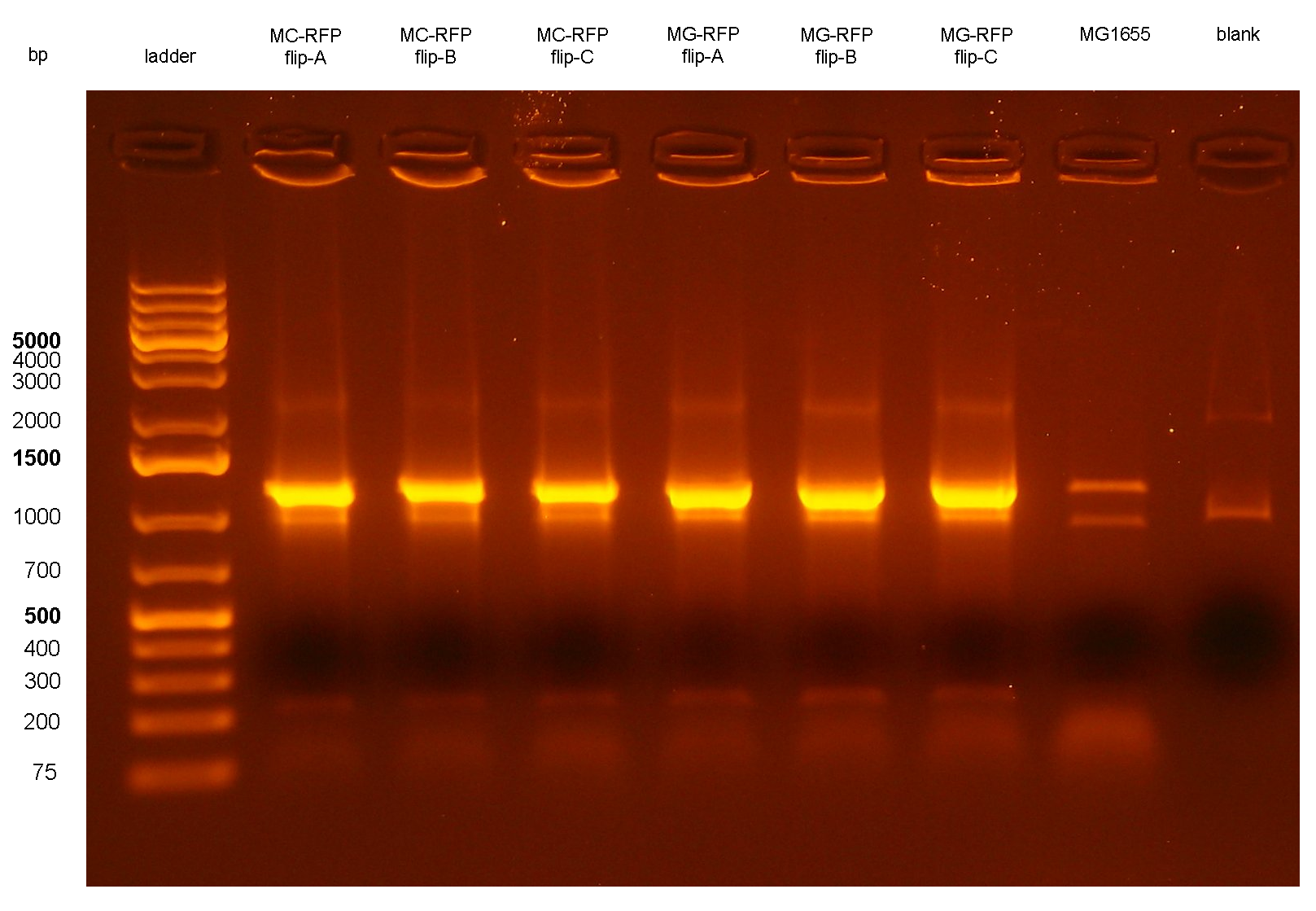
The ~2.3Kb amplicon was consistent with a single integrant of <partinfo>BBa_K300000</partinfo>-PconRFP without the R6K-CmR DNA fragment, thus validating the successful excision of the FRT-flanked DNA fragment containing R6K-CmR and confirming that PconRFP was still present in the correct locus in single copy.
All the MG-RFPflip showed a very low relative RFP synthesis rate when compared to the other strains, but the signal is systematically grater than the fluorescence of the negative control, thus validating the phenotype for the MG1655 strain. MC-RFPflip-A,B,C showed a higher fluorescence than MG-RFPflip-A,B,C. In conclusion, it has been demonstrated that, even after the marker excision process, the phenotype of the engineered cells is maintained. DiscussionA novel integrative vector for E. coli has been successfully designed, constructed and used to integrate two proof of concept protein expression systems in two commonly used E. coli strains. The results showed that the vector is fully functional and can integrate into the correct targeted locus of the host chromosome through the Phi80 site-specific recombination system by using <partinfo>BBa_J72008</partinfo>, an existing BioBrick helper plasmid from the Registry. In most cases, the integration occurs in tandem copies, probably because of the too high Chloramphenicol concentration used during the selection of integrants, which forces multiple integration of Cm-resistant constructs. This concentration was the same used during the pSC101 low copy plasmid (~5 copies per cell) selection. In some cases, it is desirable to have a single copy of the desired BioBrick in the genome, for example when the gene dosage is important. In [Haldimann A and Wanner BL, 2001] the usage of Chloramphenicol at 6 ug/ml yielded a very high percentage of single integrants. However, when tested in our lab, the MG1655 strain could survive on LB plates with Cm at 6 ug/ml and also at 8 ug/ml. For this reason a higher concentration of Cm was chosen for selection. Further studies should investigate the optimal antibiotic concentration to yield the highest single integrants percentage as possible.
Integrative standard vector for yeastMaterials and MethodsStrain: the S. cerevisiae S288C strain (<partinfo>BBa_K300979</partinfo>, genotype: MATα ρ° trp1-0) was purchased from Open Biosystems. Construction of BBa_K300001:
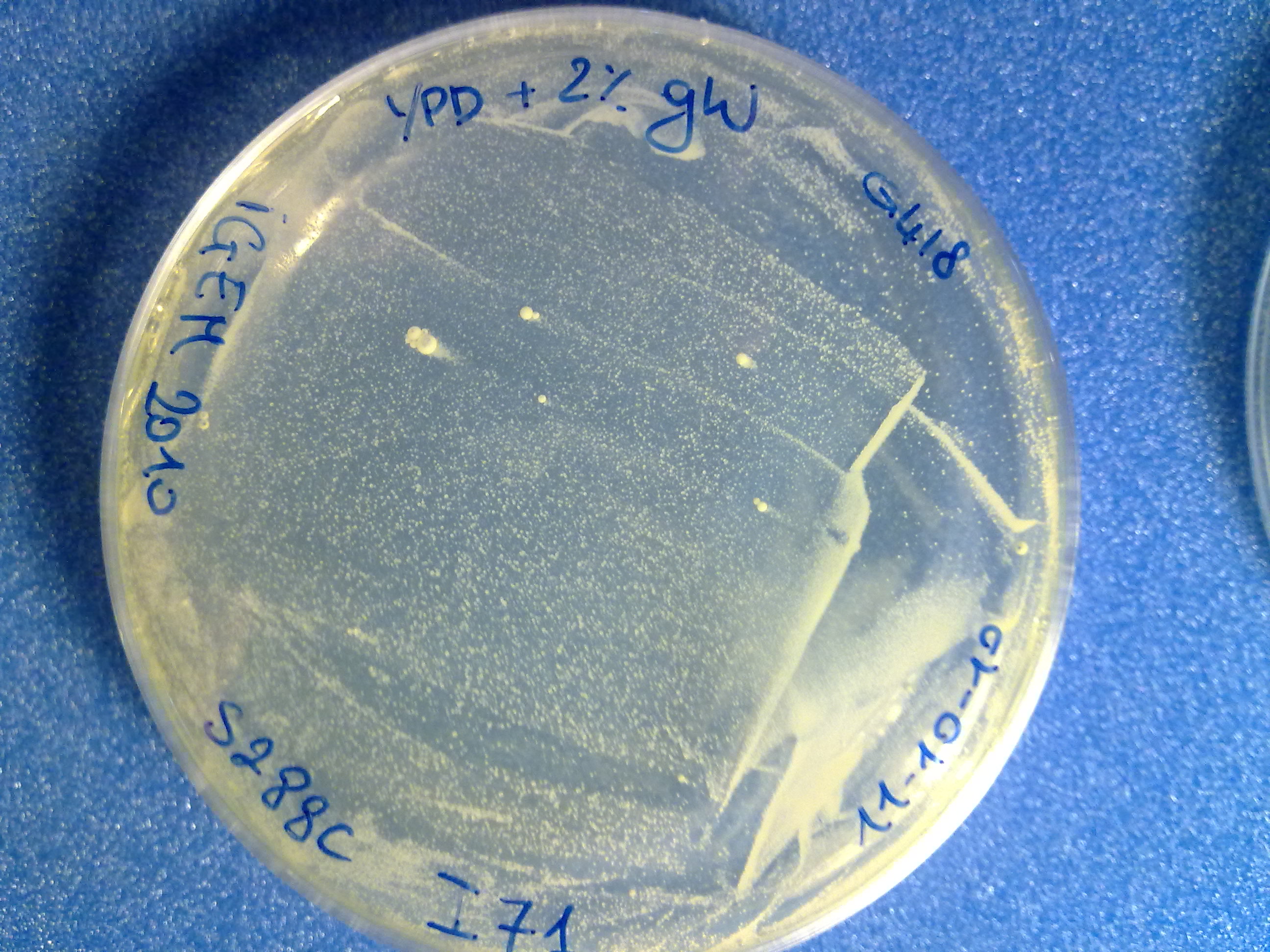
[1] http://openwetware.org/wiki/High_Efficiency_Transformation [2] Guldener U, Heck S, Fiedler T, Beinhauer J, Hegemann JH (1996), A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Research, Vol. 24, No. 13 2519–2524. ResultsThe transformed inserts and their integration efficiency in S288C are listed here:
The colony count was quite difficult (see pictures), but it is useful to have an estimation of the integration efficiency. DiscussionThe obtained results suggest that the integrative vector actually works and that the selection marker is highly specific (no colonies appeared on the "no DNA" plate). Although this integrative vector has already given promising integration results, a lot of work still remains to do:
As reported for the integrative vector for E. coli, the main information about the usage of this integrative vector for yeast is shared in the Registry. We hope to have positively contributed to the diffusion of standard biological parts for yeast, a very interesting chassis for synthetic biology for which only a limited number of parts are available in the Registry and only a few of them has been exaustively characterized.
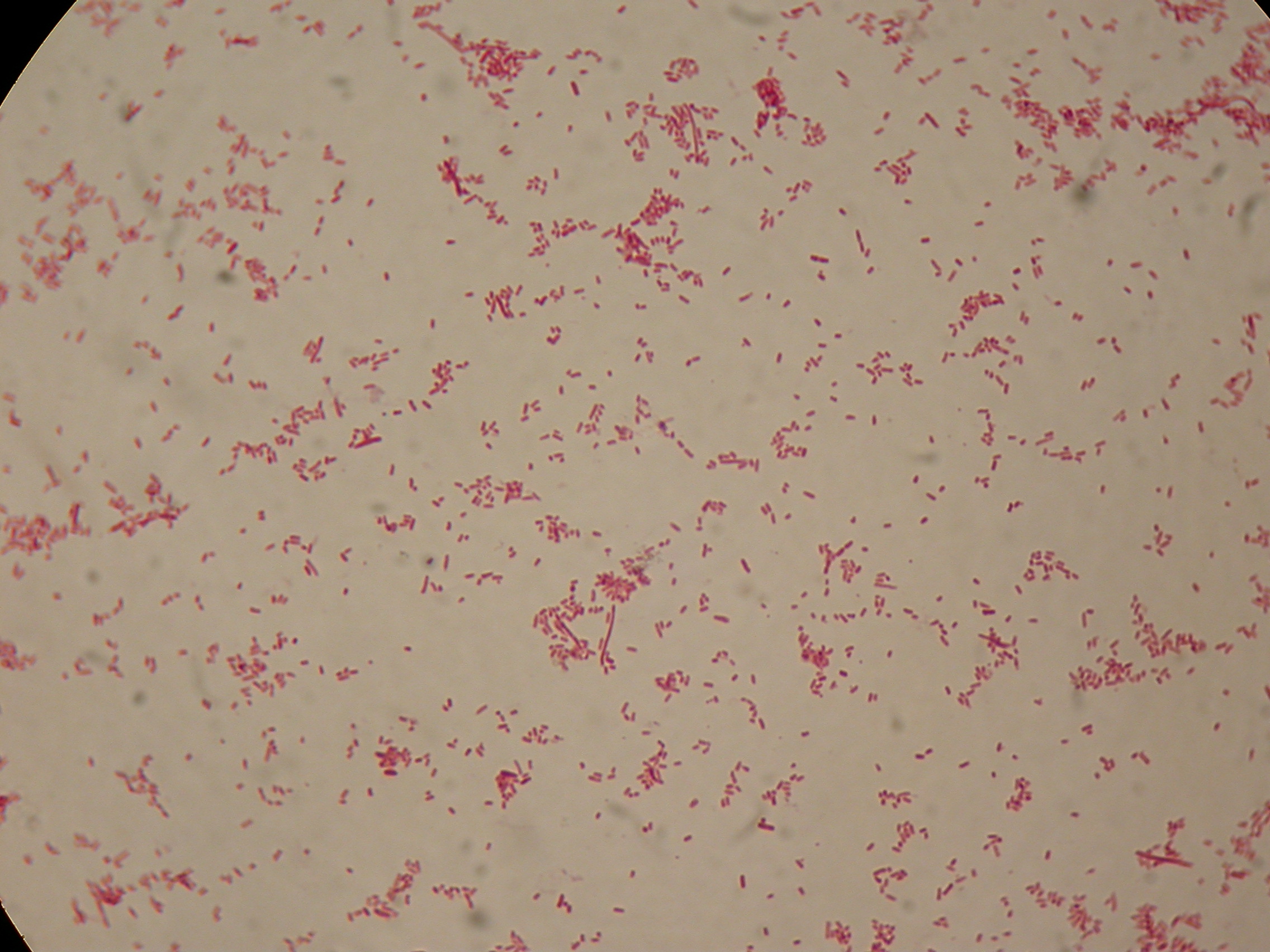
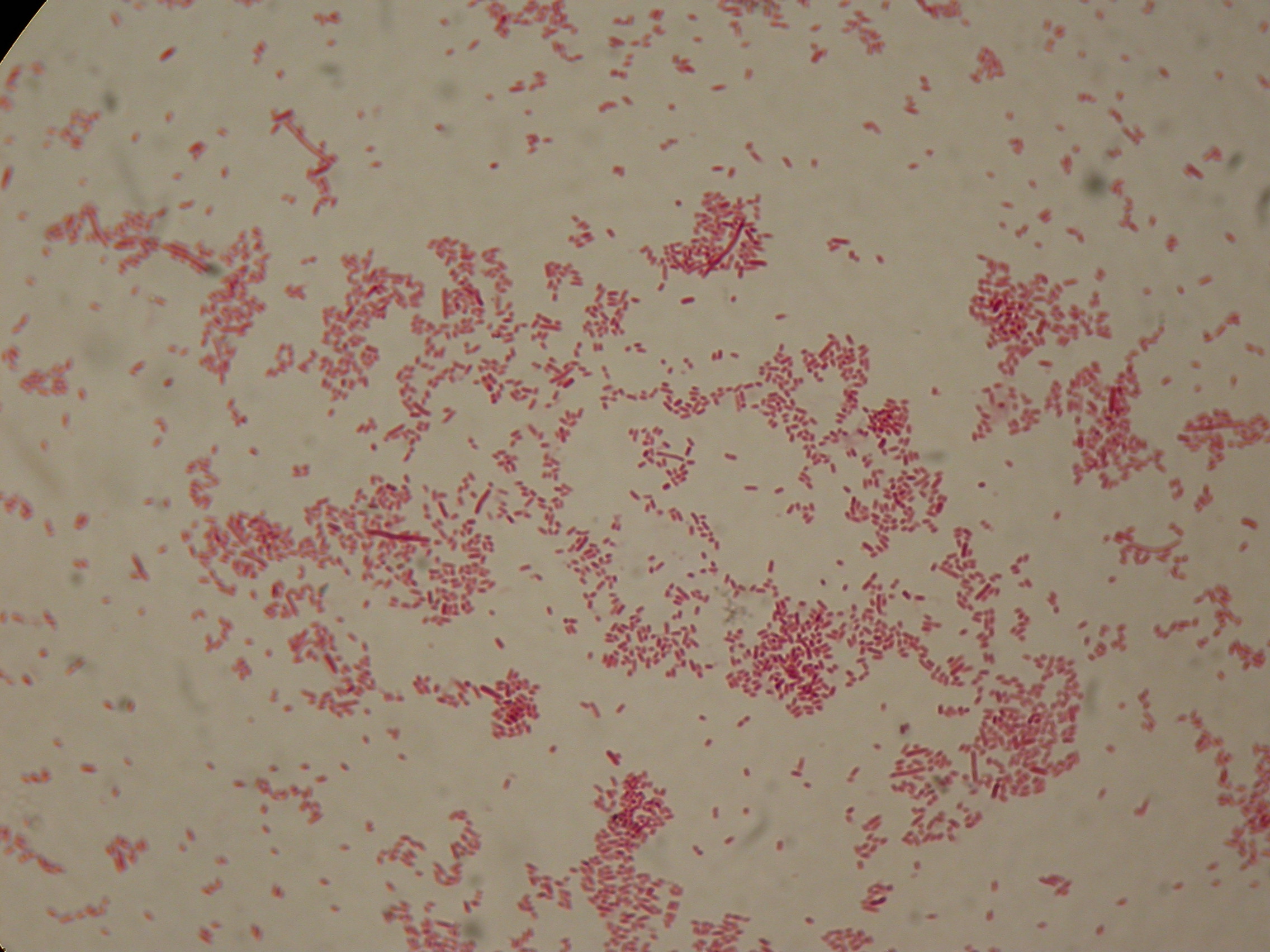
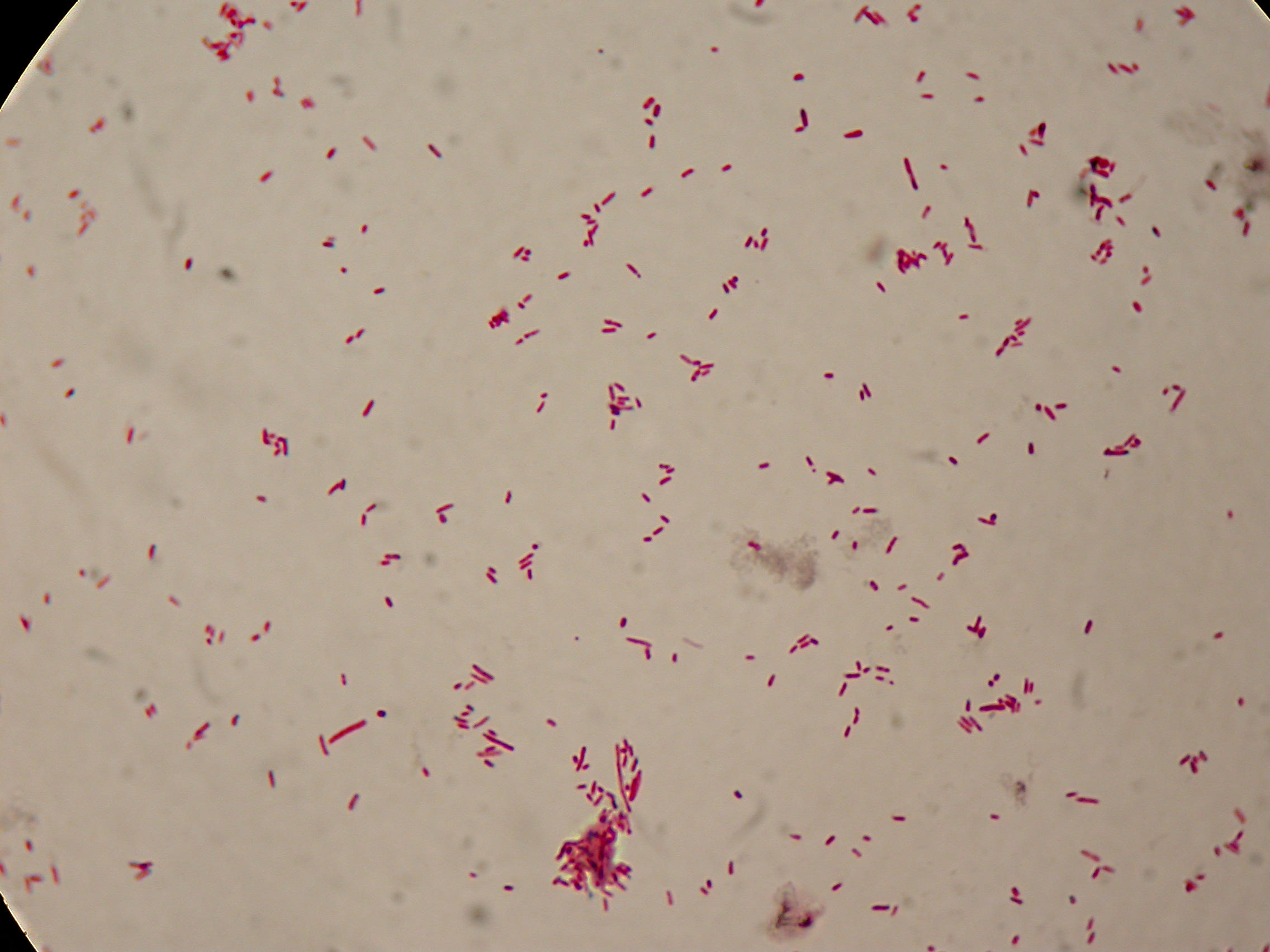
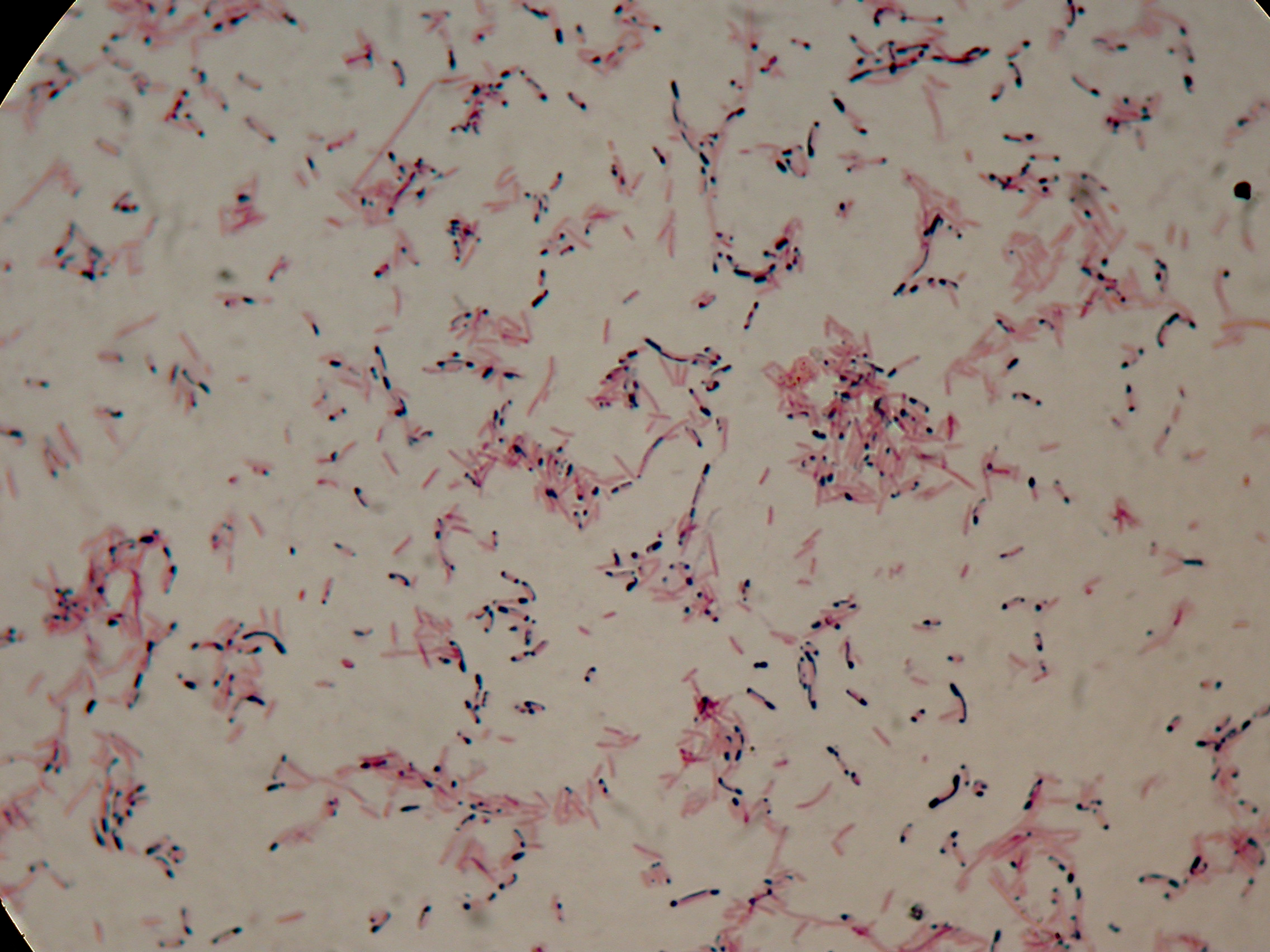
Self-cleaving affinity tags to easily purify proteinsPHB productionMethodsPreparation of samples for BioPlastic (PHB) screening:
After 5 and 30 hours' growth, Sudan Black staining protocol was performed on slides prepared for each culture. Slides were observed at the microscope. Results5 hours'Sudan Black staining protocol was performed on 70ul samples and 5 microscope slides were prepared. The resulting images are shown here: 30 hours'Sudan Black staining protocol was performed on 70ul samples and 5 microscope slides were prepared. The resulting images are shown here: DiscussionIn the above images it is clearly possible to see that after 5 hours DSMZ15372 without any kind of addition is completely similar to negative control <partinfo>BBa_B0032</partinfo> and there is no trace of bioplastic granules production. In DSMZ15372 in which IPTG or glycerol were added to the media, it is possible to see very small dark spots that could be identified as PHB granules in a few bacteria. In samples with both IPTG and glycerol, bioplastic granules are clearly visible in many cells. As expected after 30 hours negative control <partinfo>BBa_B0032</partinfo> does not show any trace of granules, while DSMZ15372 shows bioplastic granules in each experimental condition. This demonstrates that PHB can be produced without the presence of both IPTG and glycerol. In conclusion, we validated the right culture conditions for PHB production using an existing engineered strain. This enables the implementation of the purification system designed in this project. Future work can explore the possibility of producing and optimizing the PHB production with BioBrick standard parts. Fusion protein validationThe newly designed and constructed affinity tags <partinfo>BBa_K300002</partinfo>, <partinfo>BBa_K300093</partinfo>, <partinfo>BBa_K300094</partinfo>, <partinfo>BBa_K300097</partinfo> were assembled to a constitutive or inducible promoter with RBS upstream and to a Silver fusion-compatible GFP coding sequence with terminator (<partinfo>BBa_K300005</partinfo>). The resulting measurement parts were assayed to validate the bacterial growth and GFP synthesis rate of the following constructs in order to verify the right protein folding. Costitutive promoter devicesMethodsInoculum (into 5 ml LB+Amp) from glycerol stock of:
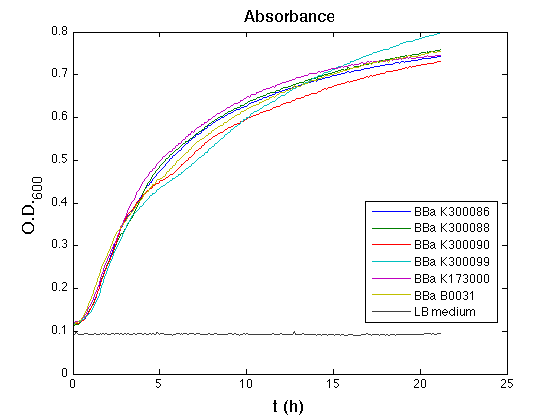
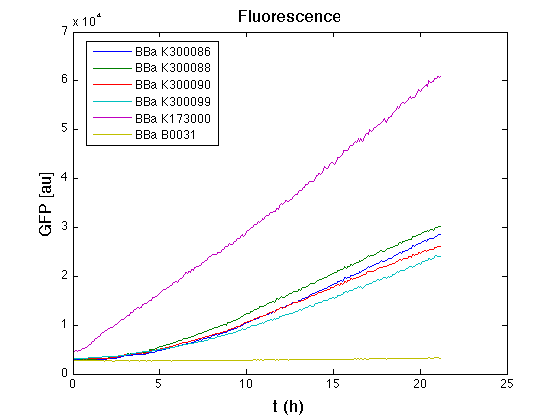
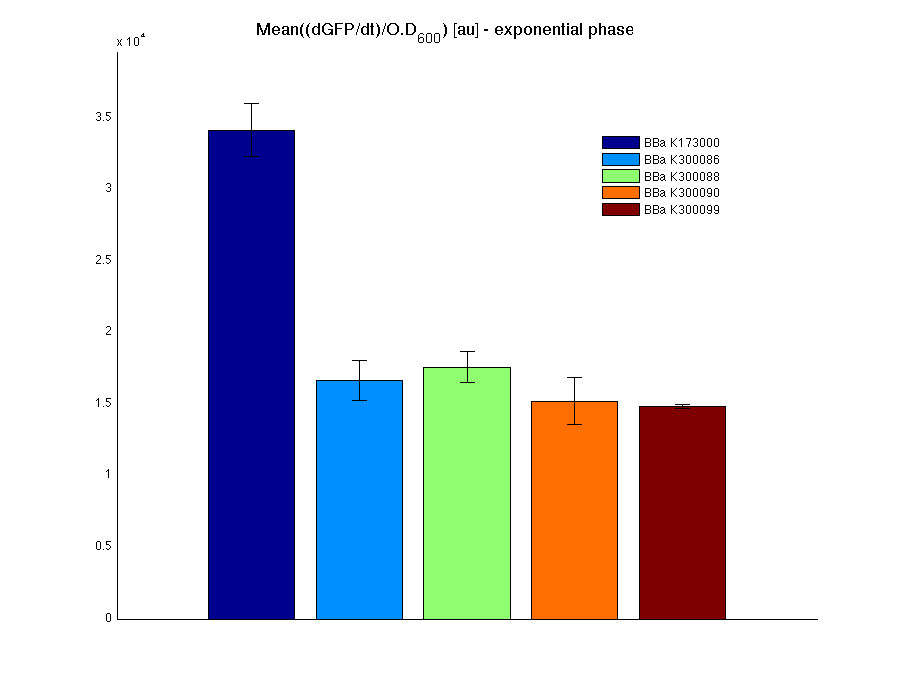
Cultures were grown ON at 37°C, 220 rpm. The following day cultures were diluted 1:100 and let grow again for about five hours at 37°C, 220 rpm. The optical density (O.D.) of each culture was than measured with TECAN Infinte F200. Samples were diluted in order to obtain the same O.D. equal to 0.02. Then we performed a 21-hour experiment with measurements of absorbance and green fluorescence every five minutes with TECAN Infinite F200; cultures were shaken for 15 seconds every five minutes. Acquired data were blanked by subtracting the media absorbance (for absorbance measurements) and the <partinfo>BBa_B0031</partinfo> fluorescence (for fluorescence measurements). Then, the relative GFP synthesis rate per cell was evaluated by computing (1/O.D.600)*dGFP/dt, where O.D.600 is the blanked absorbance of the culture of interest and GFP is its blanked fluorescence. Each value shown below is the mean of three measurements in exponential phase and error bars represent the 95% confidence interval of the mean. Results
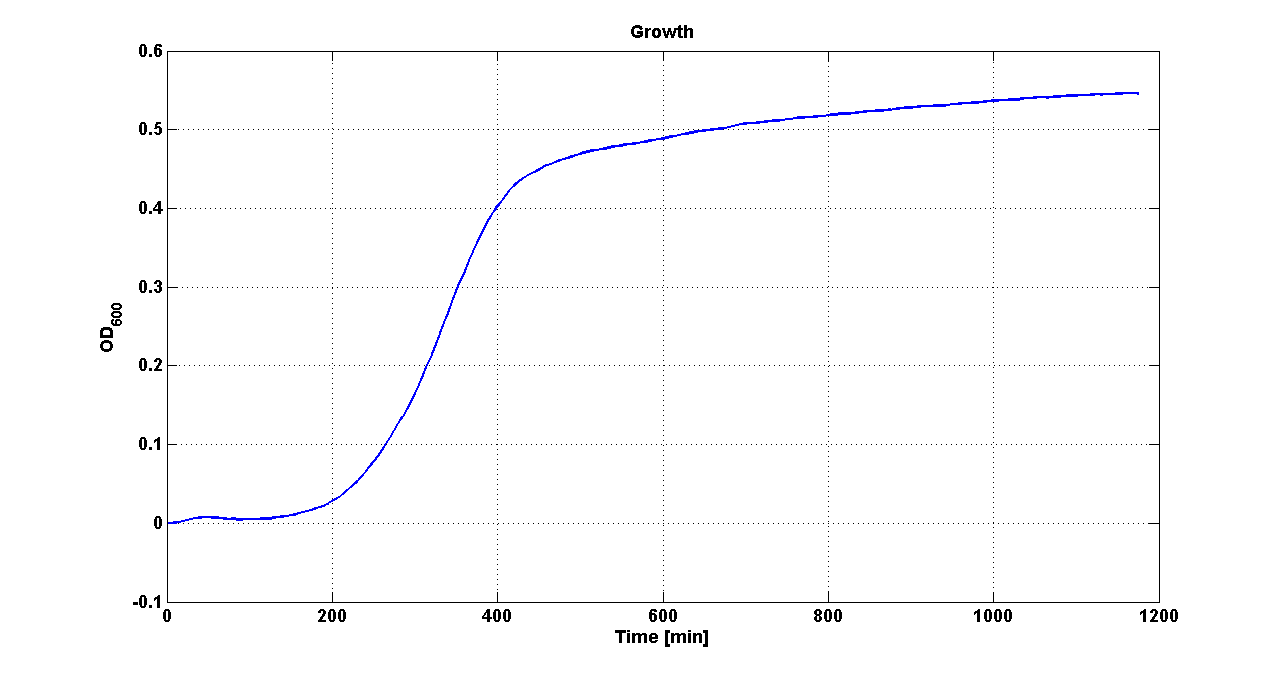
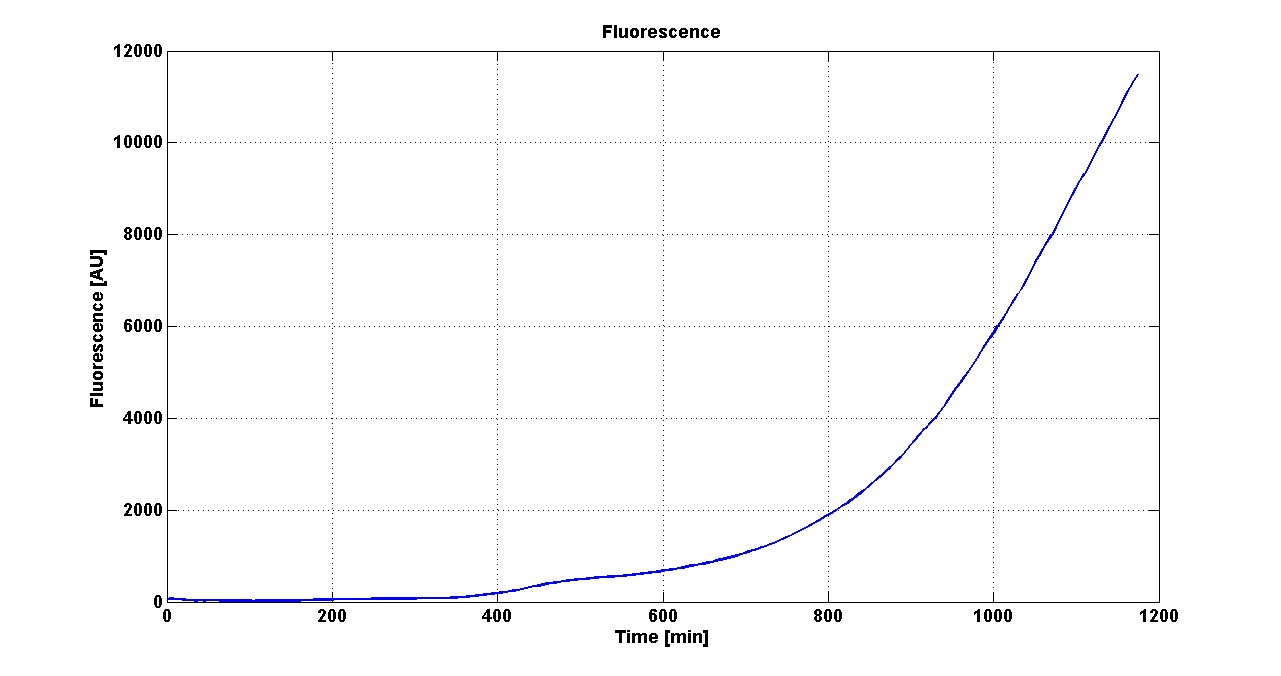
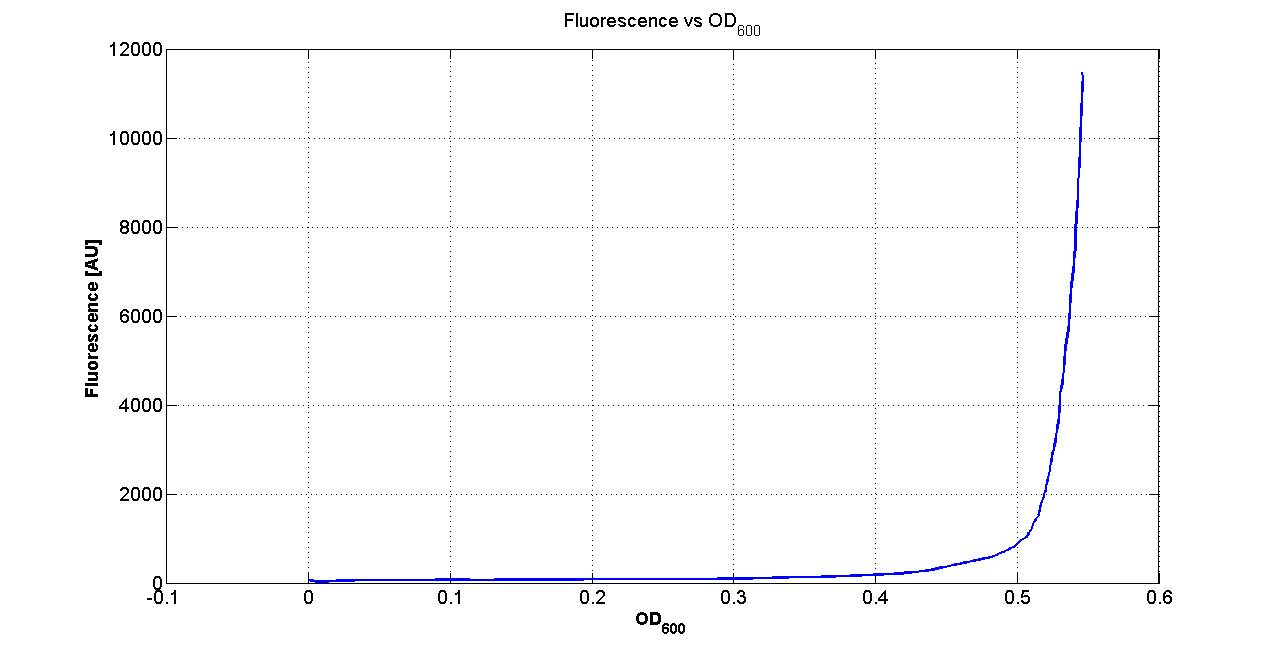
DiscussionAll the cultures showed a similar growth curve; doubling time was computed as described here in order to obtain information about the metabolic burden due to the synthesis of the studied fusion proteins. It is possible to see that all doubling times are comparable; it is possible to assert that the expression of these BioBrick parts doesn't cause abnormal stress to the cells. From GFP curve it is possible to appreciate that in <partinfo>BBa_K300086</partinfo>, <partinfo>BBa_K300088</partinfo>, <partinfo>BBa_K300090</partinfo>, <partinfo>BBa_K300099</partinfo> GFP accumulation is very similar and it is significantly different from the one of the negative control <partinfo>BBa_B0031</partinfo>. These results show that the green fluorescent protein assembled downstream of the genetic circuit is correctly folded. The mean protein synthesis rate was also computed over the exponential growth phase, showing again an appreciable GFP production rate that is about half of the positive control GFP.
3OC6HSL inducible devicesMethodsInoculum (into 5 ml LB+Amp) from glycerol stock of:
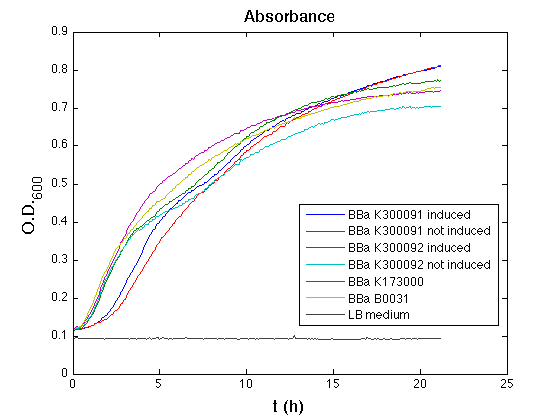
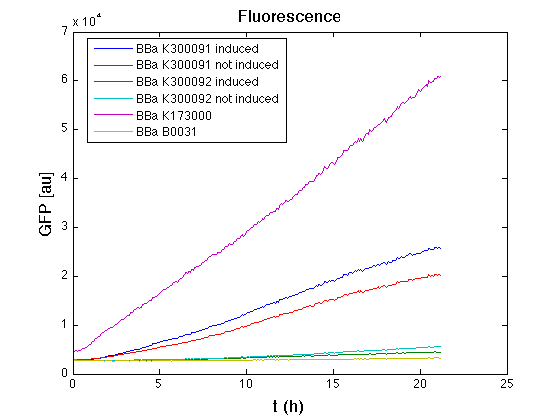
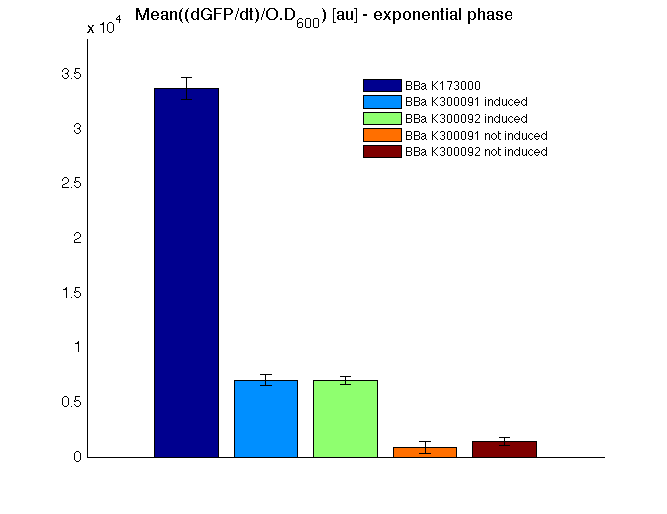
Cultures were grown ON at 37°C, 220 rpm. The following day cultures were diluted 1:100 and let grow again for about five hours at 37°C, 220 rpm. The optical density (O.D.) of each culture was than measured with TECAN Infinte F200. Samples were diluted in order to obtain the same O.D. equal to 0.02. Then we performed a 21-hour experiment with measurements of absorbance and green fluorescence every five minutes using TECAN Infinite F200; cultures were shaken for 15 seconds every five minutes. <partinfo>BBa_K300091</partinfo> and <partinfo>BBa_K300092</partinfo> constructs were induced with 100nM of HSL directly in the 96-well microplate. Acquired data were blanked by subtracting the media absorbance (for absorbance measurements) and the <partinfo>BBa_B0031</partinfo> fluorescence (for fluorescence measurements). Then, the relative GFP synthesis rate per cell was evaluated by computing (1/O.D.600)*dGFP/dt, where O.D.600 is the blanked absorbance of the culture of interest and GFP is its blanked fluorescence. Each value shown below is the mean of three measurements in exponential phase and error bars represent the 95% confidence interval of the mean. Results
DiscussionAll the cultures showed a similar growth curve; doubling time was computed as described here in order to obtain information about the burden due to the synthesis of such fusion proteins. It is possible to see that all doubling times are very similar except for induced cultures. In this case doubling time is much higher than both positive control and non-induced cultures; for this reason it is possible to assert that induction gives a high metabolic burden. From GFP curve and mean protein synthesis rate it is possible to appreciate that induced <partinfo>BBa_K300091</partinfo> and <partinfo>BBa_K300092</partinfo> GFP accumulation profiles are comparable and they significantly differ from the GFP raw time series of the negative control <partinfo>BBa_B0031</partinfo>. On the other hand not induced <partinfo>BBa_K300091</partinfo> and <partinfo>BBa_K300092</partinfo> show a profile that is very similar to the negative control. These results show that the green fluorescent protein assembled downstream of the construct is correctly folded and that the inducible system works as expected. Not induced <partinfo>BBa_K300091</partinfo> and <partinfo>BBa_K300092</partinfo> show a low GFP synthesis rate maybe due to 3OC6HSL inducible circuit leakage activity. Submitted fusion bricks for self-cleaving affinity tag constructionThe results shown above have demonstrated that the right folding can occur in a proof of concept fusion protein assembled with the newly designed synthetic affinity tags. However, the Phasin affinity with PHB granules and the self-cleavage behaviour of the Intein have not been tested yet. Several BioBrick basic and composite parts have been submitted to the Registry to enable the construction of the desired composite affinity tags for protein purification using PHB. All these parts are compatible with the Silver fusion assembly to allow in-frame protein coding sequence assembly. A list of the designed and submitted affinity tags or self-cleavable affinity tags is reported below:
Registry users are welcome to use them to create and optimize protein purification through the binding of the target protein to PHB granules, thus engineering a simple and cheap purification system. |
 "
"