Team:USTC/Project/protein/protein
From 2010.igem.org
Evelynzhang (Talk | contribs) (→= Part Ⅱ: Fusion Proteins for Transportation into BMC =) |
Evelynzhang (Talk | contribs) (→Part Ⅰ: Fusion Proteins to Identify the Localization of BMC) |
||
| Line 4: | Line 4: | ||
| - | = Part Ⅰ: Fusion Proteins to Identify the Localization of BMC = | + | ==''' Part Ⅰ: Fusion Proteins to Identify the Localization of BMC '''== |
As stated above, PduA encodes one of the seven shell proteins. In the earlier project, we have successfully linked the genes encoding the seven ''pdu'' BMC proteins by two standards and locked them into position within a vector. After transformation of this plasmid into E.coli cells, the resultant strain was grown, induced with IPTG, and incubated overnight. The cells were subsequently analyzed for the production of recombinant proteins by SDS-PAGE and the appearance of microcompartments by electron microscopy. | As stated above, PduA encodes one of the seven shell proteins. In the earlier project, we have successfully linked the genes encoding the seven ''pdu'' BMC proteins by two standards and locked them into position within a vector. After transformation of this plasmid into E.coli cells, the resultant strain was grown, induced with IPTG, and incubated overnight. The cells were subsequently analyzed for the production of recombinant proteins by SDS-PAGE and the appearance of microcompartments by electron microscopy. | ||
Revision as of 15:31, 25 October 2010
Part Ⅰ: Fusion Proteins to Identify the Localization of BMC
As stated above, PduA encodes one of the seven shell proteins. In the earlier project, we have successfully linked the genes encoding the seven pdu BMC proteins by two standards and locked them into position within a vector. After transformation of this plasmid into E.coli cells, the resultant strain was grown, induced with IPTG, and incubated overnight. The cells were subsequently analyzed for the production of recombinant proteins by SDS-PAGE and the appearance of microcompartments by electron microscopy.
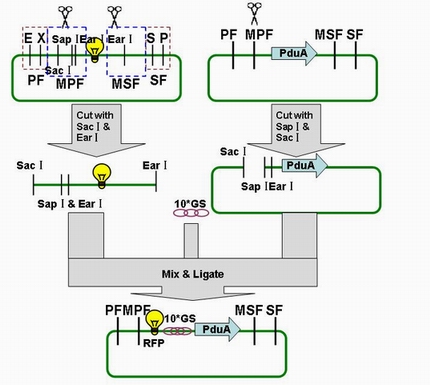
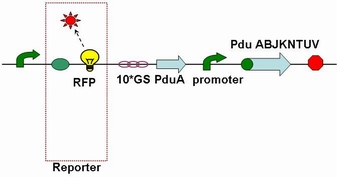
Through the fusion of GFP or RFP with the shell protein, we can easily observe the localization of BMC and distinguish whether it is right or not. The details of enzyme digestion and ligation can be seen in Fig 1a. We used pduA plasmid as a vector and RFP plasmid as an insert. Through the 4h enzyme digestion of the new standard, we got two expected segments. However, the base pairs of the two segments are not complementary. Thus, when came to the ligation, a linker (10*GS) was used. The functional fusion protein can be seen in Fig 1b.
Fig 1a: Ligation of RFP and PduA
Fig 1b: Fusion protein of RFP and PduA
If the BMC is constructed rightly, it can be detected by Western blotting and observed in fluorescence microscopy.
Part Ⅱ: Fusion Proteins for Transportation into BMC
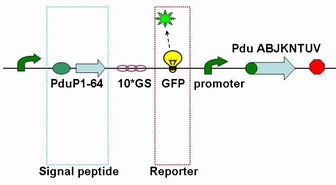
It is reported that a short N-terminal peptide is necessary and sufficient for packing enzymes into the lumen of the BMC. Fusion of the 14, 18 or 64 N-terminal amino acids from PduP to GFP or RFP resulted in their encapsulation within BMCs.
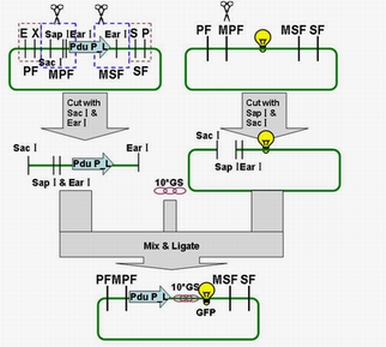
Each fusion protein (PduP[1-14]- GFP, PduP[1-18]- GFP, PduP[1-64]- GFP) was produced by the new standard. Our new standard, as it is stated above, consists of three restriction enzyme cut sites --- SacⅠ(CCTCG),EarⅠ(CTCTTC) and SapⅠ(GCTCTTC). The details of enzyme digestion and ligation can be seen in Fig 2a. We used GFP plasmid as a vector and P_L plasmid as an insert. Through the 4h enzyme digestion of the new standard, we got two expected segments. However, the base pairs of the two segments are not complementary. Thus, when came to the ligation, a linker (10*GS) was used. The functional fusion protein can be seen in Fig 2b.
Fig 2a: Ligation of Pdu P_L and GFP
Fig 2b: Fusion protein of Pdu P_L and GFP
When 14/18/64 amino acids from the N terminus of PduP were fused to GFP, the fusion protein was readily detected by Western blotting.
 "
"