Team:Michigan/Oil Sands October
From 2010.igem.org
Chillywings (Talk | contribs) (→10/23/2010) |
Chillywings (Talk | contribs) (→10/24/2010) |
||
| Line 316: | Line 316: | ||
== 10/24/2010 == | == 10/24/2010 == | ||
| + | ''Marcus and Ben'' | ||
| + | |||
| + | We re-ligated the VP130 and FimB digests that Kevin and Ben made using a 3:1 insert:vector molar ratio. We used [http://www.cynosura.org/molbiol/scripts/01_07.html this DNA calculator] to calculate the number of moles in each digest: | ||
| + | |||
| + | pSB1C3= 25 ng/uL= 18.93 fmol/uL | ||
| + | VP130= (403.24 fmol)/(50 uL)= 8.065 fmol/uL | ||
| + | FimB= (1210 fmol)/(50 uL)= 24.2 fmol/uL | ||
| + | |||
| + | We followed this [http://openwetware.org/wiki/DNA_Ligation this DNA ligation protocol]: | ||
| + | |||
| + | |||
| + | 3:1 Insert:Vector VP130 | ||
| + | *.4 uL pSB1C3 | ||
| + | *2.8 uL Vp130 | ||
| + | *1 uL 10x T4 Ligase Buffer | ||
| + | *1 uL T4 Ligase | ||
| + | *14.8 uL UP H2O | ||
| + | |||
| + | 3:1 Insert:Vector FimB | ||
| + | *.4 uL pSB1C3 | ||
| + | *.9 uL FimB | ||
| + | *1 uL 10x T4 Ligase Buffer | ||
| + | *1 uL T4 Ligase | ||
| + | *16.7 uL UP H2O | ||
| + | |||
| + | We then did the following transformations using the [[Media:Direct Plating Transformation Protocol.pdf|Direct Plating Transformation Protocol]] and the DH5alpha comp cells Nick made using his [[Media:Quick Transformation of E. coli Competant Cell Preperation.pdf| Competent Cell Preparation for Quick Transformation of E. coli]] protocol. | ||
| + | |||
| + | Transformation (Plate Resistance) | ||
| + | *Cell only control (Cm) | ||
| + | *9A pos control (Cm) | ||
| + | *9A pos control (Amp) | ||
| + | *FimB (Cm) | ||
| + | *FimB (Amp) | ||
| + | *VP130 (Cm) | ||
Revision as of 00:43, 26 October 2010
| Sunday | Monday | Tuesday | Wednesday | Thursday | Friday | Saturday | |
| Week 14 | - | - | - | - | - | 10/1/2010 | - |
| Week 15 | 10/3/2010 | 10/4/2010 | 10/5/2010 | - | 10/7/2010 | 10/8/2010 | - |
| Week 16 | - | 10/11/2010 | - | 10/13/2010 | 10/14/2010 | 10/15/2010 | 10/16/2010 |
10/1/2010
Alex
Moved broth cultures 37°C to 4°C -- 1:30pm (19-hr culture) Prepared biofilm assay plate for all pH9 tests - following Alex's biofilm assay protocol
- plate template:
- 100 μL/well: 99 μL medium + 1 μL culture
- stored plate in Lin Lab 4°C -- 2:20pm
Saved data for curves from ph7 plate Read pH7 plate ODs Performed biofilm assay on ph7 plate Started 24-hr kinetic reading for pH9 plate -- ~midnight
Discarded pH7 plate
Processed pH7 plate data
- uploaded results here
Uploaded pH7 kinetic data here
10/3/10
Alex
Ran CV biofilm assay on pH9 plate, following my protocol
- discarded plate
- uploaded results
- uploaded pH9 kinetic raw data
10/4/2010
Marcus and Ben
Performed ligation of Flu operon and pSB1AT3 according to the Ginko Bioworks Ligation protocol, with modifications.
- Calculated amount from digests to add to ligation for 1:1 and 3:1 insert:vector ratios
- Amount of DNA in digests:
- Flu PCR- 29.8 ng/uL x 17 uL= 506.6 ng / 50 uL = 10.132 ng/uL
- pSB1AT3- 240.5 ng/uL x 5 uL= 1202.5 ng / 50 uL = 24.05 ng/uL
- Used an online calculator to find moles:
- Flu PCR- 230.81 fmol/50 uL
- pSB1AT3- 537.68 fmol/50 uL
- This gives a molar ratio of 0.4293
- Amount of DNA in digests:
1:1 Ligation Mix
- 4 uL Flu PCR x 10.132 ng/uL= 40.528 ng
- 1.7 uL pSB1AT3 x 24.05 ng/uL= 40.885 ng
- 2 uL T4 Buffer
- 1 uL T4 Ligase
- 11.3 uL Ultra pure H2O
3:1 Ligation Mix
- 2 uL Flu PCR x 10.132 ng/uL= 20.264 ng
- 2.57 uL pSB1AT3 x 24.05 ng/uL= 61.809 ng
- 2 uL T4 Buffer
- 1 uL T4 Ligase
- 12.43 uL Ultra pure H2O
Note that the conversion factor for the 3:1 ligation mixture was applied in reverse, and thus it was actually a 1:3 insert:vector ratio.
Ligation program was the same as used in the Ligation protocol.
10/5/2010
Marcus
Performed transformation according to [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC145660/pdf/240536.pdf Pope & Kent (1996)].
Transformed Parts
- Flu/pSB1AT3 1:1 ligation
- Flu/pSB1AT3 1:3 ligation
- VP130
- J61100
- OmpA
- Cell only negative control
Procedure
- 2 uL of DNA was pipetted into PCR tube on ice and transferred to Lin lab
- Comp cells were thawed on ice and 50 uL was pipetted into each tube with a chilled pipette on ice
- Cells were pipetted up and down to mix throughly
- Cells were kept on ice and brought back to ERB
- Cells were plated directly onto pre-warmed plates with appropriate antibiotics
- Kevin put plates in incubator several hours before hand
It should be noted that two 200 uL aliquots of cells had to be thawed, and one was labeled "Thawed" and returned to the -80 C. Thawing should be avoided if possible.
Also, made two 5 mL LD1/LD2 co-cultures in M9+Sigma and Acros NAs, pH 7 and pH 9. Also made 2 mL pure cultures at pH 7. All culture tubes were stored in the 30 C. These cultures were made to assay growth, make new stock plates, and determine if colony morphology can be used to distinguish between species in a co-culture.
10/7/2010
Marcus
Ben moved transformation plates to 4 C yesterday, control plates were blank, and all plates except for J61100 appeared to have colonies. The Flu/pSB1AT3 ligation plates had very small colonies, so they were returned to the incubator with the J61100 plate. Also, it could be seen that many colonies on the 1:3 ligation plate appeared red, indicating that the vector had preferentially reformed. This could be because of the ratio, but dephosphorylation treatment should be used in the future for ligations.
OD600's were taken of the Pseudomonas cultures (46 hours)
- Putida pH 7- 0.157
- Fluorescens pH 7- 0.129
- Co-culture pH 7- 0.188
- Co-culture pH 9- 0.232
Surprisingly, they seem to grow better at pH 9, at least in co-culture. As expected, the co-culture did better than pure cultures. This lends some support towards Alex's biofilm assay results, however, it should still be repeated. Also, plated a serial dilution of the pH 9 co-culture, at 10-5, 10-6, and 10-7 dilutions to determine the conversion factor between OD600 and cell count. Made streak plates of all other cultures, pure cultures for stock, and pH 7 co-culture to determine if colonies could be distinguished. All cultures and plates were returned to the 30oC incubator, without shaking.
10/8/2010
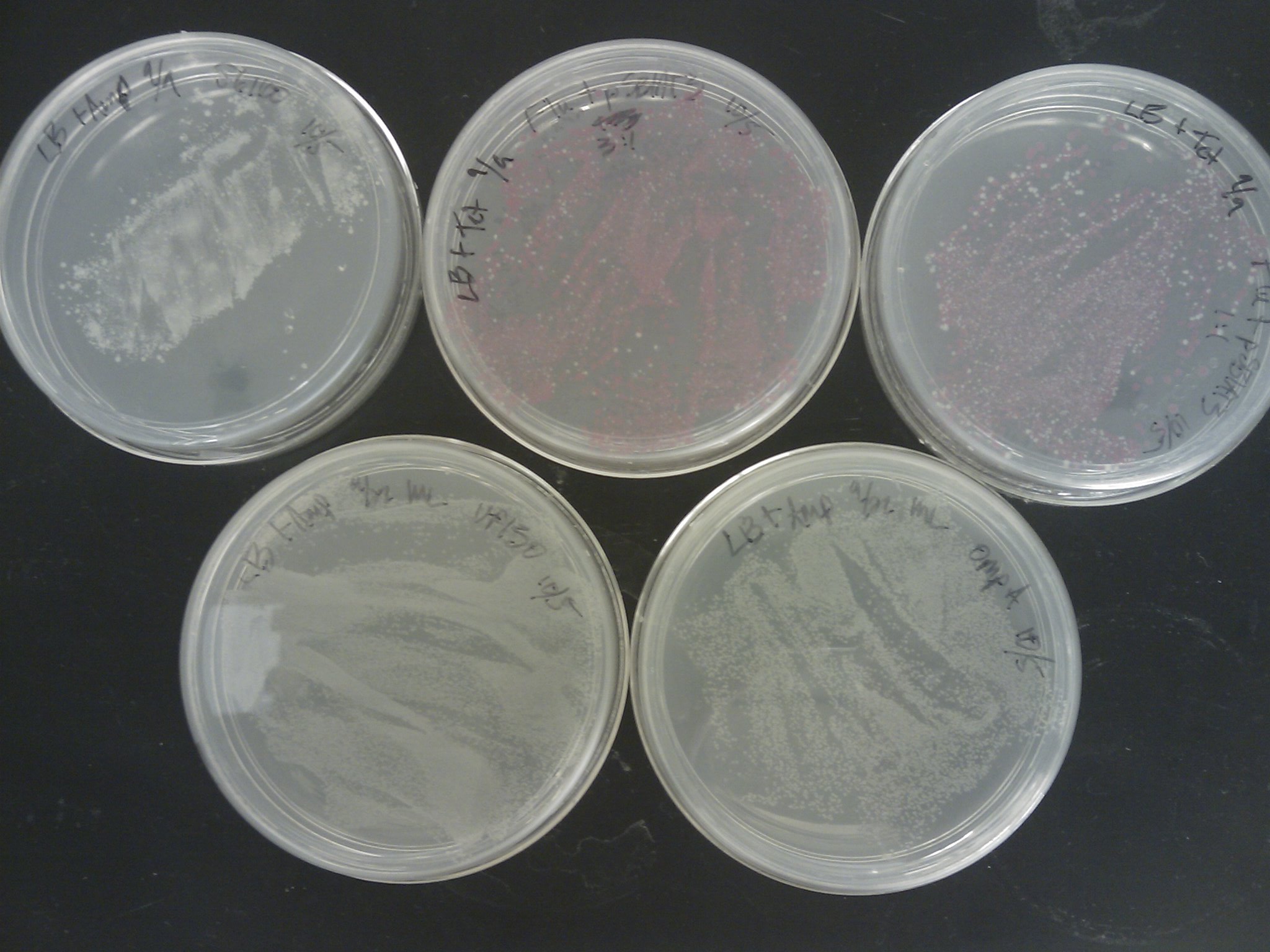
Marcus
Kevin removed the transformation plates which were placed back in the incubator to the 4oC around 1 pm.
The [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC145660/pdf/240536.pdf Pope & Kent (1996)] appears to be very efficient, as the plates were full of colonies:
Next time suspending the cells in water before spreading, or even making dilutions should be tried.
PCR Screening: 8 colonies were picked from each transformation plate except for VP130, and suspended in 50 uL of water on a microplate.
- Plate Layout
- Row 1- Flu 1:1
- Row 2- Flu 1:3
- Row 3- OmpA
- Row 4- J61100
The first two rows (Flu 1:1 and 1:3) were PCR'd by adding 1 uL of each sample to a PCR tube with a drop of mineral oil in them. A master mix was made, and 50 uL was added to each tube (should have been 49 uL).
Master Mix (18x)
- 180 uL 5x Phusion Buffer
- 18 uL 10 mM dNTPs
- 45 uL VF2
- 45 uL VR
- 9 uL Phusion polymerase
- 585 uL Ultra pure H2O
PCR Program
- 1. 95 C for 10 min
- 2. 95 C for 30 sec
- 3. 62 C for 30 sec
- 4. 72 C for 2 min
- 5. Return to step two 34 times
- 6. 72 C for 10 min
- 7. 4 C store
This PCR protocol was constructed from the following sources:
- [http://openwetware.org/wiki/Engineering_BioBrick_vectors_from_BioBrick_parts/Colony_PCR_protocol OpenWetWare BioBrick Primers Colony PCR]
- [http://openwetware.org/wiki/Endy:Colony_PCR OpenWetWare Endy Colony PCR]
- [http://www.finnzymes.com/pdf/f530sl_phusion_datasheet_2_2_low.pdf Finnzymes Phusion Manual]
Also- loosened the lids on Pseudomonas liquid cultures to provide air exchange, and returned yesterday's Pseduomonas plates to 30C incubator, without shaking.
10/11/2010
Marcus
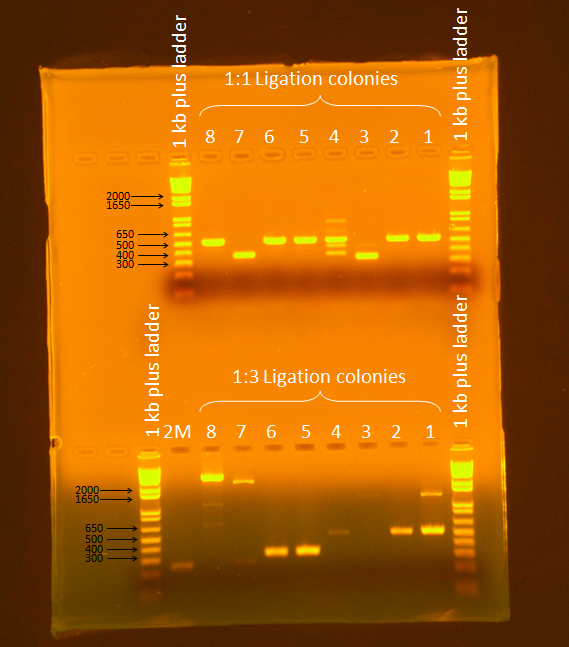
Ran a gel of PCR products from screening. Ran gel according to standard Gel Electrophoresis protocol, for 45 minutes at 86V.
Samples 7 and 8 from the 1:3 ligation appeared to be promising (expected band size= 3655 bp), so they were plated and stored in the 37C incubator overnight.
10/13/2010
Marcus
Helped Kevin with FimB PCR, discarded a few redundant plates from the refrigerator. Plates from screening were nearly lawns. Half volume (25 uL) can be plated in the future. Made overnights of single colonies from plates of samples 7 and 8, as well as VP130 from Nick's electroporation and my direct plating for a miniprep tomorrow.
10/14/2010
Marcus
Overnights made were only 2 mL, and should've been 5 mL. So 3 mL of fresh LB (with antibiotics) was added to 50 mL tubes, and the cultures were added to the fresh LB at 1 pm. Minipreps were made according to the MODIFIED Miniprep at room temperature and stored in the 4C.
10/15/2010
Marcus and Ben
Nanodropped minipreps from yesterday, the prep from sample 8 appears to be a dud.
pSB1AT3-Flu #7- 69.86 ng/uL pSB1AT3-Flu #8- 16.45 ng/uL VP130 HS- 96.88 ng/uL VP130 EP- 99.34 ng/uL
Also, showed Ben and Natalie how to make plates. We made 10 Amp and 10 Tet plates.
Digested VP130 HS according to DNA Digest and [http://ginkgobioworks.com/support/BioBrick_Assembly_Manual.pdf Ginko BioWorks BioBrick Assembly Manual].
Digest Mix:
- 5.2 uL VP130 HS miniprep
- 37.3 uL Ultrapure water
- 0.5 uL BSA
- 5 uL NEB Buffer 2
- 1 uL EcoRI
- 1 uL PstI
Incubation:
- 37C for 15 min
- 80C for 20 min
Ligated according [http://www.roche-applied-science.com/pack-insert/4898117a.pdf Roche Rapid Ligation Protocol].
Ligation Mix:
- 1 uL pSB1C3 (linearized from Registry)
- 7 uL VP130 HS digest
- 2 uL 5x DNA Dilution Buffer
- 10 uL 2x T4 Ligase
Incubation:
- Room temperature fro 5 min
Transformed according to Direct Plating Transformation Protocol, but made 1:1 and 1:3 dilutions by adding 50 uL of cold, sterile DI water to the comp cells and mixing before each plating.
10/16/2010
Nick
10-16-10 Psuedomonas Competent Cell Preparation
Protocol - Competent Cell Preparation for Quick Transformation of E. coli
Ben Parker started the overnight cultures and assisted in OD measurement and initial centrifugation steps.
A colony of P. putida and P. fluorescence was picked from LB agar plates (10/6/10) and grown up in an overnight culture in 2ml Amp at 30 C.
The overnight culture was added to 100 ml of LB media and grown at 30C for 4 hours.
Protocol was carried out approximately 20 minutes after the following OD600 was recorded:
Blank - water
P. putida – 0.587
P. Fluorescence – 0.610
One centrifuge tube containing p. putida culture failed during centrifugation at 5000 rpm. The protocol was altered by reducing the speed to 2500 rpm after this point to reduce the chance of failure. The cells are stored in the 4C fridge in room 1239.
10/17/2010
Marcus and Ben
Nanodropped Nick's miniprep of pSB1AT3-Flu (#8) from yesterday, but found low concentrations of about 13-18 ng/uL since each mL of overnight was miniprepped individually. The samples were pooled by using a repurification protocol in the miniprep kit, eluted in Ultrapure water, and got a concentration of 40.2 ng/uL. The sequencing core requires 50 ng/uL, so a 5 mL overnight of pSB1AT3-Flu was made for Ben and Nick to miniprep tomorrow, and a 2 mL overnight of DH5alpha for comp cells.
10/18/2010
Kevin and Ben
Miniprepped incubated overnight containingn pSB1AT3. Note that during step 3 or 4 the supernatent was vortexed. DNA eluded with EB buffer. DNA was digested using EcoR1 and Xbal, then ligated using a 1 hour protocol. It was then combined with bacteria. It was diluted by 1/2 and plated, then diluted to 1/4 and plated again on tet plates.
10/19/2010
Marcus
Made 10 Tet and 10 Cm plates. Also, Nick and I transformed DH5alpha comp cells from -80 iGEM box with extra pSB1AT3-J23100-Flu ligation Ben and Kevin made yesterday. Transformed according to Direct Plating Transformation Protocol. We also transformed P. putida and P. fluorescens with pSB1AT3 as a test. No dilutions were made. It was noted while plating the cells that the Pseudomonas strains somehow wound up with less than 50 uL of cells. And due to the cells flocculating during storage, the Pseudomonas strains also had be vortexed to resuspend the cells.
10/21/2010
Marcus
Some colonies were found on the pSB1AT3-J23100-Flu transformation plates that Ben and Kevin made. Colonies were PCR'd following the PCR Screening Protocol.
- Made 18x master mix, and screened 16 colonies (8 from each plate, 1:1 and 1:3)
- Primers were dissolved using the following amounts to make 50 uM stocks:
- VF2- 33.9 nmol/.05 nmol/uL= 678 uL
- VR- 40 nmol/ .05 nmol/uL= 800 uL
- Used a 2 min extend
Also, put the Cm and Tet plates from Tuesday in the 4C.
10/23/2010
Marcus
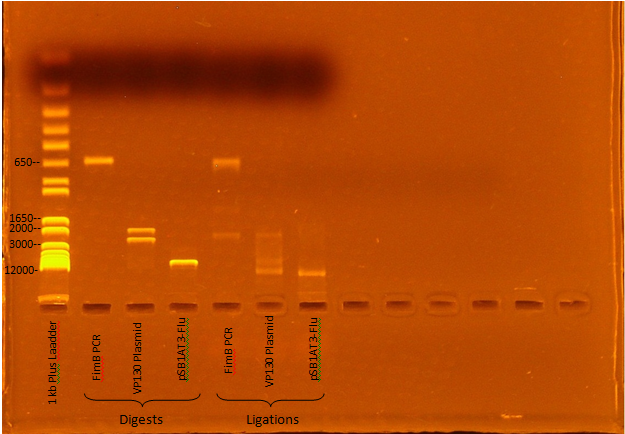
Jeremy M. ran a gel of Kevin and Ben's digest and ligation mixtures:
Expected Digest bands
- FimB- 638 bp
- VP130- 2376, 1925 bp
- pSB1AT3-Flu- 6770, 15 bp
The digests appear to have worked, although the ligations seem to have been incomplete (Roche Rapid Dephos & Ligation Kit).
10/24/2010
Marcus and Ben
We re-ligated the VP130 and FimB digests that Kevin and Ben made using a 3:1 insert:vector molar ratio. We used [http://www.cynosura.org/molbiol/scripts/01_07.html this DNA calculator] to calculate the number of moles in each digest:
pSB1C3= 25 ng/uL= 18.93 fmol/uL VP130= (403.24 fmol)/(50 uL)= 8.065 fmol/uL FimB= (1210 fmol)/(50 uL)= 24.2 fmol/uL
We followed this [http://openwetware.org/wiki/DNA_Ligation this DNA ligation protocol]:
3:1 Insert:Vector VP130
- .4 uL pSB1C3
- 2.8 uL Vp130
- 1 uL 10x T4 Ligase Buffer
- 1 uL T4 Ligase
- 14.8 uL UP H2O
3:1 Insert:Vector FimB
- .4 uL pSB1C3
- .9 uL FimB
- 1 uL 10x T4 Ligase Buffer
- 1 uL T4 Ligase
- 16.7 uL UP H2O
We then did the following transformations using the Direct Plating Transformation Protocol and the DH5alpha comp cells Nick made using his Competent Cell Preparation for Quick Transformation of E. coli protocol.
Transformation (Plate Resistance)
- Cell only control (Cm)
- 9A pos control (Cm)
- 9A pos control (Amp)
- FimB (Cm)
- FimB (Amp)
- VP130 (Cm)
 "
"