Team:Harvard/allergy/notebook
From 2010.igem.org
(→For Allergen panel) |
(→For pKannabil and pHannibal) |
||
| Line 469: | Line 469: | ||
Extension Time: 30 sec | Extension Time: 30 sec | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
''Concentrations'': PDK:46.5 ng/uL & 36.5 ng/uL | ''Concentrations'': PDK:46.5 ng/uL & 36.5 ng/uL | ||
Revision as of 19:41, 24 October 2010

notebook
| Week 1 | 06-14-2010 | 06-15-2010 | 06-16-2010 | 06-17-2010 | 06-18-2010 |
| Week 2 | 06-21-2010 | 06-22-2010 | 06-23-2010 | 06-24-2010 | 06-25-2010 |
| Week 3 | 06-28-2010 | 06-29-2010 | 06-30-2010 | 07-01-2010 | 07-02-2010 |
| Week 4 | - | 07-06-2010 | 07-07-2010 | 07-08-2010 | 07-09-2010 |
| Week 5 | 07-12-2010 | 07-13-2010 | 07-14-2010 | 07-15-2010 | 07-16-2010 |
| Week 6 | 07-19-2010 | 07-20-2010 | 07-21-2010 | 07-22-2010 | 07-23-2010 |
| Week 7 | 07-26-2010 | 07-27-2010 | 07-28-2010 | 07-29-2010 | 07-30-2010 |
| Week 8 | - | 08-03-2010 | 08-04-2010 | 08-05-2010 | 08-06-2010 |
| Week 9 | 08-09-2010 | 08-10-2010 | 08-11-2010 | 08-12-2010 | 08-13-2010 |
06-14-2010 [ top ]
06-15-2010 [ top ]
06-16-2010 [ top ]
06-17-2010 [ top ]
06-18-2010 [ top ]
Progress
- Identified allergens in arabidopsis, strawberries,tomatoes, and carrots and created primers for amplifying out these allergens (see primers and sequences)]
- Extracted cDNA (rna extraction followed by rt-pcr) from strawberries and tried to PCR out our strawberry allergen (Fra a 1), but were unble ato do so
- Looked to find introns ~ 800 bp to use to create our hairpins since it was difficult to acquire pHannibal vector w/ the pdk intron
Future Directions
- Look towards ordering amiRNA for silencing
- Waiting on arabidopsis plants to grow to extract genomic DNA (mon)
- Look to find introns to use in creation of hairpin DNA
- Cloning (digesting/ligating our sense/antisense/intron sequences necessary for hairpin creation into one vector to create biobricks)
06-21-2010 [ top ]
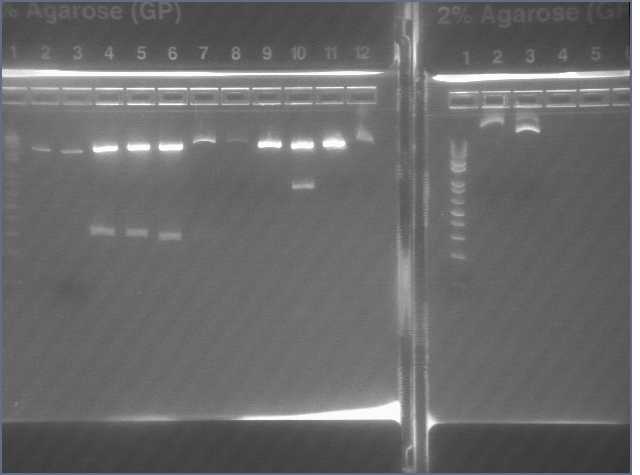
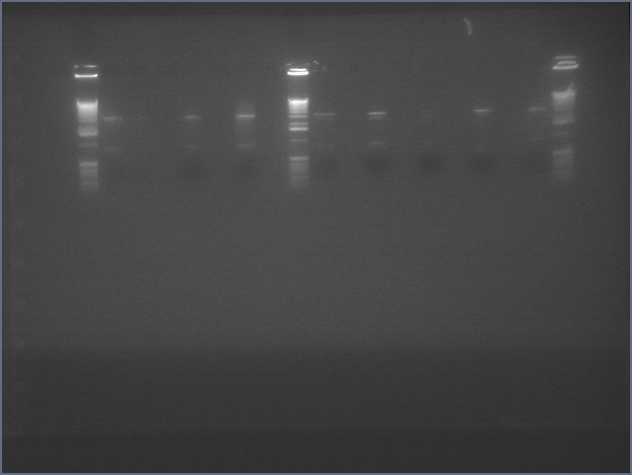
- PCR amplification of gDNA from Arabadopsis thaliana for sense and antisense parts of LTP, Bet v 1, and Ger3.
- Diagnostic Digest of PCR products
Results
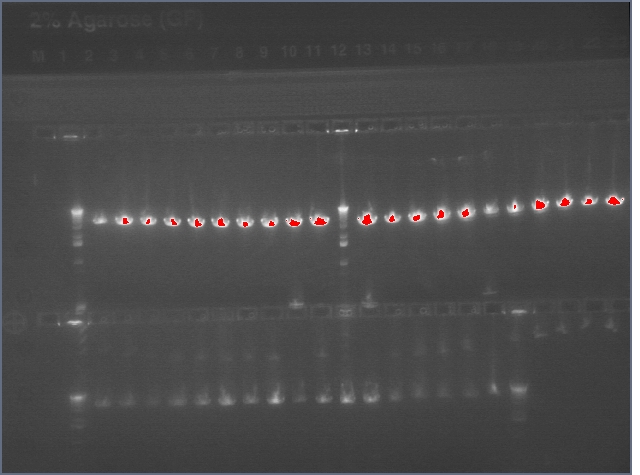
Lanes:
2. LTP sense
11. Ger 3 sense
12. Ger 3 antisense
06-22-2010 [ top ]
- Gel electrophoresis of PCR products
- Gel digest and purification of LTP sense, Ger sense and Ger antisense
- Digest with Xba and Pst
- Purification of parts
Results
Gel purification yielded 2.6, 7.8, and 7.9 ng/μL of LTP sense, Ger sense, and Ger antisense parts. We were able to obtain 1.5, 4.3 and 0.5 ng/μL of digested LTP sense, Ger 3 sense, and Ger 3 antisense, respectively, from the digest purification.
06-23-2010 [ top ]
- Digest and purification of vector V0120
- Ligation of LTP sense, Ger sense, and Ger antisense parts into V0120
Results
Ligations were plated, but no viable colonies grew.
06-24-2010 [ top ]
- PCR of arabadopsis allergens for remaining parts
- PCR purification
- Diagnostic gel electrophoresis of PCR products
Results
We successfully obtained the rest of the allergen sense and antisense parts. Nanodrop of the PCR products showed the following concentrations in ng/μL
| Gene | PCR Products | Purified DNA |
|---|---|---|
| LTP 1 sense | 220.9 | 20.4 |
| LTP 1 antisense | 62.6 | 12.2 |
| Bet v1.1 sense | 216.2 | 15.4 |
| Bet v1.1 antisense | 228.1 | 11.5 |
| Bet v1.2 sense | 205.5 | 9.2 |
| Bet v1.2 antisense | 68.5 | 7.7 |
| Ger 3 sense | 73.3 | 11.2 |
| Ger 3 antisense | 21.0 | 8.5 |
Diagnostic gel electrophoresis of a small sample of the PCR products showed proper length bands (300 bps) for all the parts.
06-25-2010 [ top ]
- Digest Arabadopsis allergen panel with Xba1 and Pst1
- Purify digested DNA
- Digest vector V0120 (from Team Vector)
- Purify digested V0120
Results
Stored digests at -20 degrees Celsius for ligations tomorrow.
06-28-2010 [ top ]
- Create amiRNA
- PCR reaction of RS300 plasmid with primers containing RNAi sequence for Arabadopsis allergens (see methods for detailed protocol for working with RS300)
- Diagnostic gel electrophoresis
- PCR purification to obtain parts
- Ligate parts together
- Three in one sewing PCR primed with A and B
- Two in one sewing PCR primed with A and II, followed by another two in one PCR reaction primed with A and B.
Results
9 reactions total for creating amiRNA parts. We used the following temperatures for the different primers and a 15 seconds extension time.
Annealing Temp: (LTP & Bet)60C (GFP)69C
The concentrations of parts 1, 2, and 3 of each allergen are:
LTP 1: 5.4 ng/uL 2: 5.5 ng/uL 3: 1.2 ng/uL GFP 1: 17 ng/uL 2: 6.6 ng/uL 3: 15.1 ng/uL Bet 1:7.4 ng/uL 2: 29.6 ng/uL 3: 34.3 ng/uL
Gel electrophoresis of the parts showed strong bands for each individual part, and correct bands for sewing ligations of all three allergens.
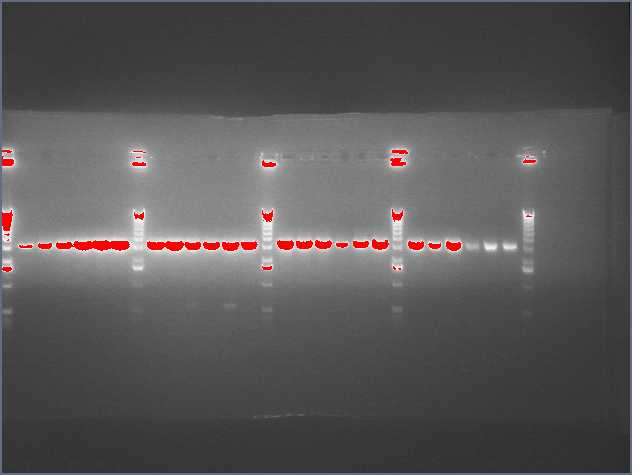
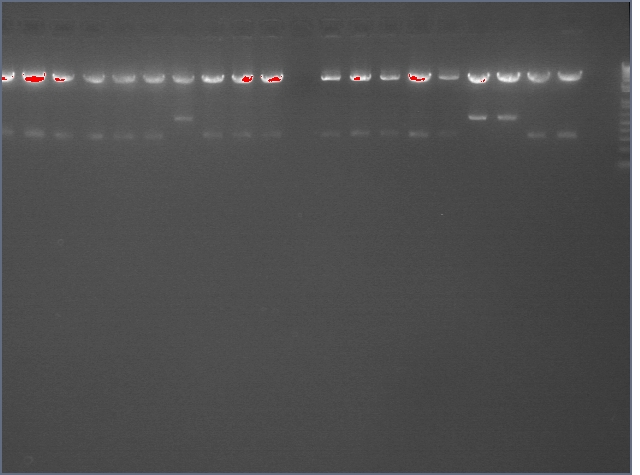
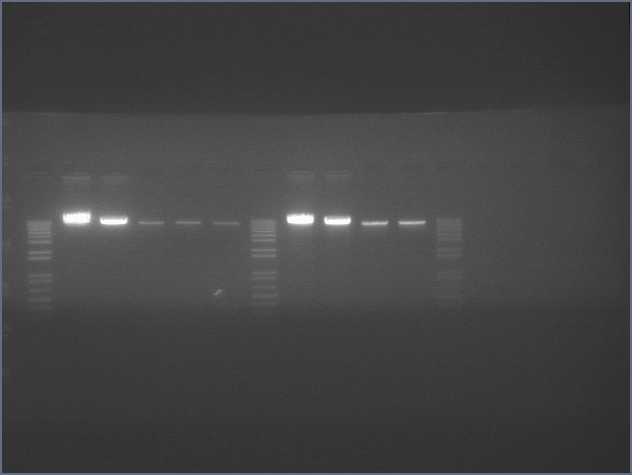
The image below show results sewing PCR reactions. Lanes are:
1 (ladder); 2(LTP 1+2); 3 (Bet 1+2); 4(GFP 1+2); 5(Ladder); 6(LTP 1,2,3); 7(Be 1,2,3); 8(GFP 1,2,3); 9(LTP 1,2,3(using .5ul)); 10(GFP 1,2,3 (using .5 uL)). Lanes 2-4 should be ~a little less than 450 bp and 6-10 should be ~450bp
06-29-2010 [ top ]
Today, we did not do work with hpRNA and focused our efforts on amiRNA. We obtained undigested V0120 with a death gene insert from Team Flavor, and we have inserts from previous PCR reactions that all need to be digested and ligated into V0120.
Procedures:
- Digestion of Allergen Panel and V0120 with Xba and Pst
- Ligation of Allergen inserts into Vector Backbone
- Transformation of E. Coli
Results
Digestion of allergens yielded the following concentrations:
Bet: 58.2 ng/uL; LTP (.5): 38.6 ng/uL; GFP (.5): 43.9 ng/uL; GFP: 29.1 ng/uL; LTP: 60.4 ng/uL; Backbone: 20.2 ng/uL
06-30-2010 [ top ]
- Ligations for full amiRNA and V0120 (w/o death gene)
note: .5 denotes multistep pcr reaction from yesterday in which 0.5 uL of part 3 instead of 1.0 uL of gfp/ltp were use in one of the three-in-one assemblies of parts 1,2,3 w/ primers A&B
Ligation Reactions
- Used 50 ng of backbone and 150 ng of insert (ratio of insert to vector ~.14
- Did 1:10 dilutions for the digested inserts
- PCR of PME3 and PAL2
- saw no bands from last night's pcr so we re ran it
Annealing Temp: 69C Extension Time: 30 sec
2 PCR reactions (PME & PAL)
Concentrations: PME: 250 ng/uL PAL: 134 ng/uL
- gel: lane 1:ladder; lane 2: Pal3; lane 3: Pme 2; lane 4: ladder
- Sent amiRNA Bet for sequencing
- Ligation of Antisense/sense allergens into V0120 (lacking death gene)
- Used 34 ng of backbone and 10 ng of insert (ratio of insert to vector ~.09375
07-01-2010 [ top ]
- Ligations for full amiRNA and V0120 (w/o death gene)
note: .5 denotes multistep pcr reaction from yesterday in which .5 uL of part 3 of gfp/ltp were use in one of the giant assemblies of parts 1,2,3 w/ primers A&B
Ligation Reactions
- Used 50 ng of backbone and 150 ng of insert (ratio of insert to vector ~.14
- Did 1:10 dilutions for the digested inserts
- PCR of PME3 and PAL2
- saw no bands from last night's pcr so we re ran it
Annealing Temp: 69C Extension Time: 30 sec
2 PCR reactions (PME & PAL)
- Concentrations: PME: 250 ng/uL PAL: 134 ng/uL
- gel: lane 1:ladder; lane 2: Pal3; lane 3: Pme 2; lane 4: ladder
- Sent amiRNA Bet for sequencing
- Ligation of Antisense/sense allergens into V0120 (lacking death gene)
- Used 34 ng of backbone and 10 ng of insert (ratio of insert to vector ~.09375
07-02-2010 [ top ]
- Check on sequences for Bet
- Digest Pme3 and Pal 2 introns/Ligate w/ V0120/Transform
- PCR purified digests
- Concentrations: Pme (ranges from 20-36 ng/uL); Pal (ranges from 20 to 28 ng/uL)
- Ran backbone on gel and gel purified (Concentration: 3.1 ng/uL)
- PCR purified digests
- Ligations of Pme/Pal w/ V0120
- bbone 2 was potentially contaminated at a higher concentration 96.1 ng/uL
- Check on transformations of amiRNA (5 transformations) and antisense/sense allergens (8 transformations)
- There were colonies from the amiRNA transformations
- We grew up 24 colonies (3 for each GFP, GFP .5, LTP, LTP .5, BET) ~ 10 am to be ready ~3 pm for minipreps)
- The other 8 transformations did not show any colonies--we put it back in to see if we would get colonies (only 100 uL instead of 200 uL was plated)
- To do: Grow up colonies to miniprep, run on gel, sequence
- There were colonies from the amiRNA transformations
- pHannibal and pKannibal arrived today (hannibal is amp resistant and kannibal is kan resistant)
- We transformed these plasmids
07-06-2010 [ top ]
Team Allergy
Today, we isolated plasmid that contained antisense and sense sequences of the allergen panel and introns PME and PAL that we grew last week.
Procedures
For amiRNA
- Nanodrop of miniprepped amiRNA plasmids
- Diagnostic digest of amiRNA plasmids
- E Gel of digested amiRNA
- Send plasmids to Genewiz for sequencing
For ihpRNA
- Miniprep of plasmids containing introns for hpRNA
- Nanodrop of miniprepped intron plasmids
- Miniprep of plasmids containing Allergen Panel sense and antisense
- Nanodrop of minipreped allergen plasmids
- Diagnostic Digest of introns and allergens
- E Gel of digested parts
- Extract pKannibal and pHannibal vectors and transform into E. coli
- PCR of the PDK intron in pKannibal and pHannibal
Results
For amiRNA
Miniprep of turbo bacteria for plasmids containing sequences for amiRNA
Should obtain plasmids containing the complete sequence for amiRNA. The resulting concentration should be relatively high (<100ng/μL) because all growing bacteria would have the plasmids inside.
Nanodrop of miniprepped amiRNA plasmids
We used 2μL of DNA per nanodrop.
| Sample # --> | One | Two | Three |
|---|---|---|---|
| GFP | 134.4 | 167.6 | 207.2 |
| GFP 0.5 | 163.3 | 106.9 | 230.9 |
| LTP | 134.0 | 127.8 | 165.5 |
| LTP 0.5 | 158.5 | 137.2 | 113.5 |
| Bet | 129.4 | 78.9 | 124.4 |
Diagnostic digest of amiRNA plasmids Digested 200~500ng of plasmids with EcoR1 and Spe1. We will then run the digested vectors on an E Gel and look for DNA that is the length of the insert. If such DNA appears on the gel, then we will send it off to be sequenced.
E Gel of digested amiRNA
We had to run 21 samples, so we used two gels. The orders were:
Gel #1
- 1kb Ladder
- GFP 1
- GFP 2
- GFP3
- GFP 0.5 1
- GFP 0.5 2
- GFP 0.5 3
- LTP 1
- LTP 2
- LTP 3
- LTP 0.5 1 no visible band for neither plasmid nor insert
- LTP 0.5 2
Gel #2
- 1kb Ladder
- LTP 0.5 3
- Bet 1
- Bet 2
- Bet 3
- Neg Control for hpRNA (digested with Xba and Pst, and ligated without new inserts) no visible insert band
- Neg Control for intron (digested with Eco and Spe, and ligated without new inserts)
- PME Good no visible insert band
- PME Dubious no visible insert band
- PAL Good colony 1 no visible insert band
- PAL Good colony 2
- 1kb Ladder
For ihpRNA
Miniprep of plasmids containing introns for hpRNA Nanodrop of miniprepped intron plasmids
We used 2μL of DNA per nanodrop.
| Insert Name | Good | Dubious | Colony 1 | Colony 2 | Colony 3 |
|---|---|---|---|---|---|
| PME | 85.6 | 66.8 | |||
| PAL | 67.7 | 108.8 | |||
| Neg Control, Intron | 74.0 | ||||
| Neg Contron, hpRNA | 63.5 | 30.1 | 37.3 |
- Miniprep of plasmids containing Allergen Panel sense and antisense (from overnight cultures)--still need to do preps of cultures grown today
- Nanodrop of minipreped allergen plasmids
- Diagnostic Digest of introns and allergens
- E Gel
Gel #1
Lane 1: 1kb Ladder
Lanes 2-4: Ger Sense
Lanes 5-7: Ger Antisense
Lanes 8-10: LTP Sense
Lanes 11-13: LTP Antisense
Lanes 14-16: Bet 2 Sense
Lanes 17-19: Bet 2 Antisense
Lanes 20-22: Bet 1 Sense
Lanes 23-25: Bet 1 Antisense
phannibal and pkannibal
Extracted vectors from a piece of paper per instructions. Transformed vectors into E. coli. Miniprep and PCR to extract PDK intron.
- pcr to get out pdk intron
07-07-2010 [ top ]
Yesterday, we ended by transforming pKannabil and pHannibal into E. coli and running a diagnostic digest of digested allergen panel. Yesterday's plates did not grow, and the diagnostic digest did not have good results.
- Today, we will be redoing diagnostic digests of the allergen panel plasmids to see if we can see our desired bands (300 bp and 3200) and re-transforming pKannabil and pHannibal.
- If the digested allergen panel runs correctly on a gel, then we will send the plasmids off for sequencing.
- If nothing gel does not run correctly, then we will go back to re-digest V0120 with a death gene insert instead of YFP and ligate allergen inserts, transform, miniprep, and see if we get the correct insert. **If some run correctly and some did not, we will collect more samples from our culture plates for the allergens that did not work and redo the digests until we find a sample with the correct plasmid.
- If the pKannabil and pHannibal plates grow, then we will miniprep to purify the plasmids containing the PDK intron, PCR the intron, and purify the intron.
- Today, we are also running the PDK intron obtained yesterday to see if it is the correct length.
First thing tomorrow, we will culture E. coli containing plasmids for our allergen panel to obtain more plasmids.
Procedures
For pKannabil and pHannibal:
- Electronic Gel Electrophoresis of PDK introns (from PCR reaction yesterday, PCR off DNA from the eluted vectors)
- Re-grow E. coli containing pKannabil and pHannibal plasmids
- Culture colonies
- Miniprep colonies
- PCR for PDK intron
- Diagnostic Gel to check for correct length
- Repeat PCR of PDK intron using different Tm (gradient pcr)
For Allergen panel
- Digest of Plasmids containing allergen inserts
- Diagnostic Gel
- Collect additional samples for allergens that did not work
Results
For pKannabil and pHannibal
Transformation of pHannibal pKannibal
This transformation we performed slightly differently from the one yesterday. This protocol is nicknamed the "quick and dirty transform." The transformation we started yesterday of pH and pK didn't work (no growth on either plate).
Quick and Dirty Transform:
- Mix
- 2ul 10x {pHannibal, pKannibal}
- 20ul Turbo E. Coli
- 220ul SOC Broth
- Plate on {LB+(pHannibal->LB+Amp, pKannibal->LB+Kan), LB} Agar, grow
PCR of PDK intron
- PCR of PME3 and PAL2
- saw no bands from last night's pcr so we re ran it
Annealing Temp: 69C Extension Time: 30 sec
Concentrations: PDK:46.5 ng/uL & 36.5 ng/uL
Didn't work. Our theory is that the Tm we used (the one IDT sent us) was too high. IDT assumes that the entire primer is going to anneal to a strand of DNA when they calculate their Tm, but since half of our primer won't anneal (the half that doesn't anneal was used to prepend/postpend the biobrick sequences) the Tm IDT calculates will be too high. Here are more correct calculations from Primer3:
OLIGO start len tm gc% any 3' seq LEFT PRIMER 4229 20 51.11 30.00 8.00 3.00 CCAATTGGTAAGGAAATAAT RIGHT PRIMER 5021 20 61.95 45.00 6.00 0.00 TTCGAACCCAATTTCCCAAC
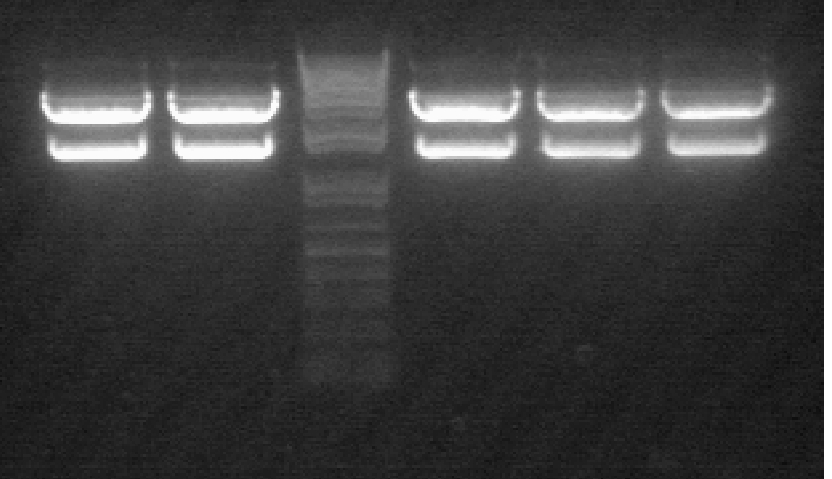
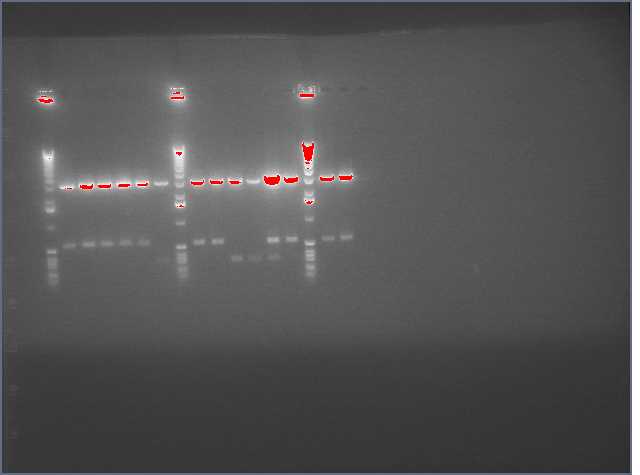
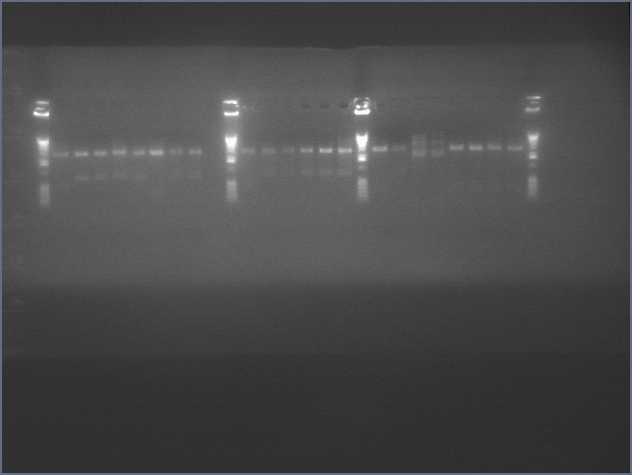
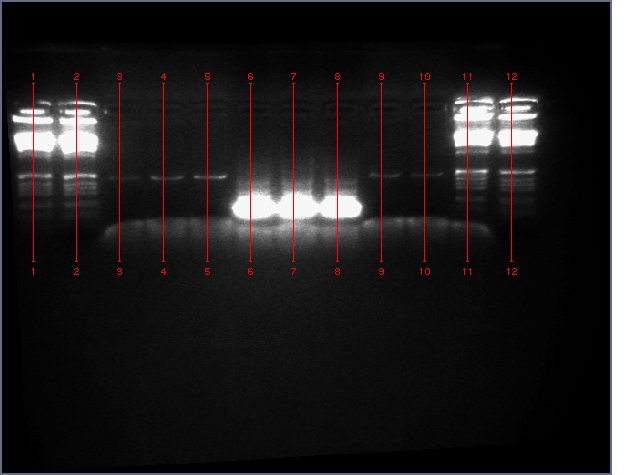
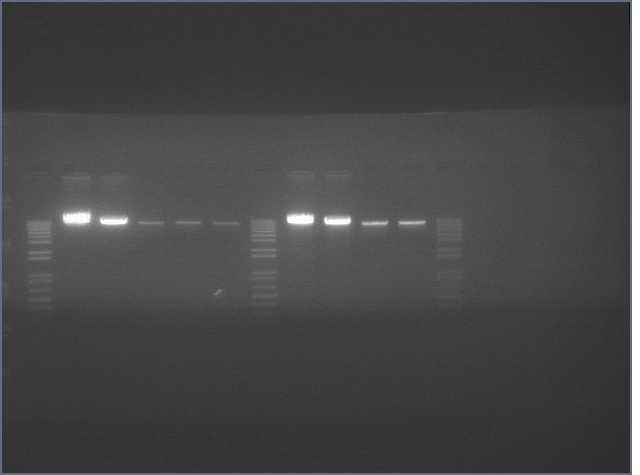
See Gel Image Below: ladder, lane 3 and 5 are pdk intron
Lanes:
3. pdk intron
5. pdk intron
Gradient PCR
Annealing Temps: 50,55,60,65
Extension Time: 30 sec
8 reactions
| Name | Amount (uL) |
|---|---|
| 10mM dNTPs | 1 |
| 5x Phusion Buffer | 10 |
| Phusion Polymerase | .5 |
| Arabadopsis Genomic DNA | 7 ng |
| Fwd/Rev Primer | .5 uM each (1uL 50x) |
| Water | 35.5 |
Concentrations: PDK: 12.7 ng/uL
Remaining steps
- Culture colonies
- Miniprep colonies
- PCR for PDK intron
- Diagnostic Gel to check for correct length
- Repeat PCR of PDK intron using different Tm.
For Allergen panel
Digest of Plasmids containing allergen inserts Digested with Xba and Pst using Fast Fermentas digest protocol
Out of the 24 lanes, 6 were successful. Of the 8 allergens, 3 were successful. Samples of LTP sense, Betv1 sense, and Ger sense had one band at around 3kbps and one band at 300bps. The other genes had one band at 3kbps and additional longer bands on the scale of twice or three times the length of the plasmid.
For the allergens that did not work, we will go back to our bacteria plates and sample 5 additional colonies for each allergen, grow up 10 mL overnight cultures, and miniprep so that we can redo the diagnostic digest. We are hoping that a larger sample size would allow us to find colonies with the correct plasmid.
Remaining Steps
- Collect additional samples for allergens that did not work
- Culture them
- Miniprep
- Diagnostic Digest and Gel
07-08-2010 [ top ]
amiRNA
- Sent out plasmids for sequencing w/ new primers
Allergen Panel
- Sent out for sequencing this morning using new primers
- Minipreps of Allergen Panel
- Concentrations:
LTP 1,2,3: 265.5 ng/uL, 507.1 ng/uL, 275.5 ng/uL;
GerA 1,2,3,4: 283.1 ng/uL, 185.9 ng/uL, 182.4 ng/uL, 314.7 ng/uL;
Bet 2A 1,2,3,4: 246.6 ng/uL, 251.4ng/uL, 550.3 ng/uL, 235.8 ng/uL;
Bet 2S 1,2,3,4,5,6: 613 ng/uL, 296.8 ng/uL, 333.1 ng/uL, 467.3 ng/uL, 230.3 ng/uL, 129.4 ng/uL;
Bet 1A 1,2,3,4,5: 505.8 ng/uL, 632.1 ng/uL, 561.7 ng/uL, 594.4 ng/uL, 609.2 ng/uL
- Diagnostic Digest
Gradient PCR of PDK intron
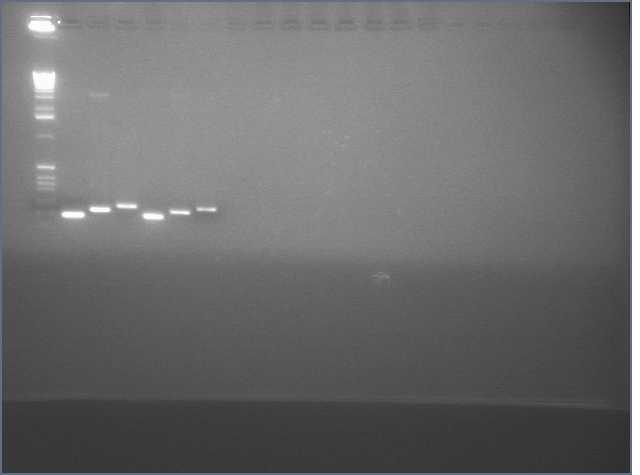
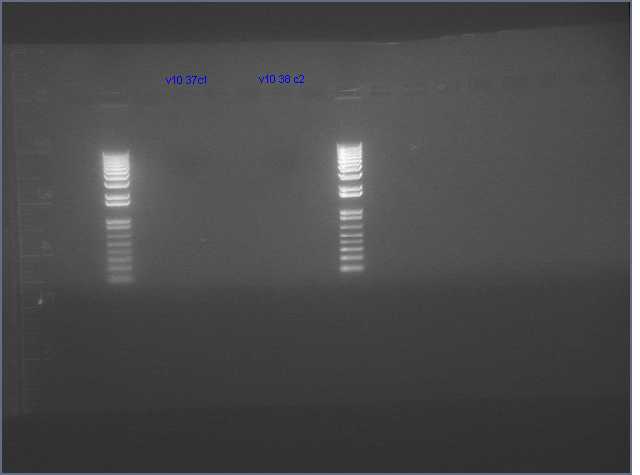
Gel of gradient PCR of the 741bp PDK intron out of pHANNIBAL and pKANNIBAL. Lanes are (left to right, temperatures are annealing temperatures used in the gradient PCR) 1kb+ ladder, pHAN 50ºC, pKAN 50ºC, pHAN 55ºC, pKAN 55ºC, pHAN 60ºC, pKAN 60ºC, pHAN 65ºC, pKAN 65ºC.
It appears that pHANNIBAL worked at 50, 55ºC but pKANNIBAL didn't.
Concentration: hannibal pdk 12.7 ng/uL kannibal pdk: 15.6 ng/uL
Did a PCR of the PCR product
Annealing Temps: hannibal pdk (50 C); kannibal pdk (55 C)
Extension Time: 30 sec
12 reactions (6 hannibal pdk; 6 kannibal pdk)
| Name | Amount (uL) |
|---|---|
| 10mM dNTPs | 1 |
| 5x Phusion Buffer | 10 |
| Phusion Polymerase | .5 |
| pdk | 1 uL DNA (12.6 and 16.5 ng) |
| Fwd/Rev Primer | .5 uM each (1uL 50x) at .5 uM |
| Water | 35.5 |
- Growing up 5 mL cultures of pHannibal and pKannibal for getting more DNA for pdk pcr
- Gel purifying/digesting/ligating/transforming pdk intron into e.coli
07-09-2010 [ top ]
- PCRd pHAN.pdk, pKAN.pdk (they're the same 741bp pdk) using the values that worked in the gradient, got them in high ODs. Put them in box a2 rows 3-4.
| Temp(ºC) | Source | Label | Concentrations |
|---|---|---|---|
| 55 | H | HPDK55 | 64.0,190.9,66.4 |
| 50 | H | HPDK50 | 119.3,125.5,92.4 |
| 55 | K | KPDK55 | 84.3,97.3,106.5 |
| 50 | K | KPDK50 | 52.5,93.5,71.0 |
07-12-2010 [ top ]
Today, we received sequences for the allergen , intron, and amiRNA inserts. Since 4 of the 8 allergen inserts were not correctly done (and were not set for sequencing) and we want to repeat the inserts that DID work, we will redo ligations for the entire panel.
Procedures
- Check Sequencing Results
- Redo Ligations for allergen Panel
Results
Sequencing Results
- amiRNA
- GFP #1 & GFP.5 #3 worked
- Bet,LTP did not work
- intron
- Pal(#2) matched
- ihpRNA
- LTP Sense 1,2,3 and Ger Sense 1 matched
- Bet sense did not match
Full report (annotated sequences): File:Report.zip
Redo Ligations w/ phosphatized backbone for entire allergen panel,
PME did not grow, we will not use it since we have good PDK and PAL introns.
PAL and PDK will be digested and inserted into a plasmid containing a sense strand of an allergen.
Add PDK to amiRNA vectors if sequences match up
07-13-2010 [ top ]
Digestion
| pLTPS1 | pLTPS2 | pLTPS3 | GerS1 | ||
|---|---|---|---|---|---|
| DNA | 10 | 8 | 7 | 10 | |
| FD Buffer (10x) | 2 | 2 | 2 | 2 | |
| diH2O | 6 | 8 | 9 | 6 | |
| Spe | 1 | 1 | 1 | 1 | |
| Pst | 1 | 1 | 1 | 1 |
- add phosphatase to backbone (15 min)
| Pal2 | PDK | |
|---|---|---|
| DNA (~ 1 ug) | 9 | |
| FD Buffer (10x) | 2 | 2 |
| diH2O | 7 | |
| Xba1 | 1 | 1 |
| Pst1 | 1 | 1 |
- gel purify backbones LTPS: 8.7 ng/uL Ger: 9.4 ng/uL
- pcr purify digested pdk: PDK1 14.4 ng/uL; PDK2 22.4 ng/uL; PDK3 11.4 ng/uL; PDK4 15.3 ng/uL
Ligations
Ligations of PDK w/ V0120 (lacking YFP)
| Pdk1 | Pdk2 | Pdk3 | Pdk4 | |
|---|---|---|---|---|
| DNA Insert | 2.36 | 2.98 | 1.52 | 2.22 |
| T4 DNA ligase buffer (10x) | 2 | 2 | 2 | 2 |
| diH2O | 12.64 | 12.02 | 13.48 | 12.78 |
| T4 DNA ligase | 1 | 1 | 1 | 1 |
| DNA Backbone (~50ng) | 2uL | 2uL | 2uL | 2uL |
Ligations of PDK/Pal into V0120 + Sense Allergens (LTP & GER)
| PDK1 | PDK2 | PDK 3 | PDK 4 | ||
|---|---|---|---|---|---|
| DNA Insert (~36ng/ ~32 ng)) | 2.51; 2.27 | 1.62;1.46 | 3.18; 2.86 | 2.37;2.13 | |
| T4 DNA ligase buffer (10x) | 2 | 2 | 2 | 2 | |
| diH2O | 8.49; 9.73 | 9.38; 10.54 | 7.82;9.14 | 8.63;9.87 | |
| T4 DNA ligase | 1 | 1 | 1 | 1 | |
| DNA Backbone (~50ng/(~47ng) | LTP1S 6; GerS 5 | LTP1S 6; GerS 5 | LTP1S 6; GerS 5 | LTP1S 6; GerS 5 |
07-14-2010 [ top ]
- Genomic DNA extraction
- PCR for allergen panel from genomic DNA
- Grew up cultures of transformations from yesterday (LTPS+intron; GerS+intron; PDK+V0120)--tubes weren't cloudy so we grew overnight cultures again
8 reactions (Ger S/A; BetS/A; Bet1.2 S/A; LTP S/A)
Annealing Temp: 65 C Extension Temp: 30 sec
| Name | Amount (uL) |
|---|---|
| 10mM dNTPs | 1 |
| 5x Phusion Buffer | 10 |
| Phusion Polymerase | .5 |
| DNA | 1 uL |
| AB primer mix (1x) | 2.5 |
| Water | 35 |
Concentrations: BetS: 109.1 ng/uL; BetA: 90.5 ng/uL; GerS: 69 ng/uL; GerA: 46 ng/uL; LTPS:36.7 ng/uL; LTPA:5.2 ng/uL; Bet1.2S: 77.4 ng/uL; Bet1.2A:66.2 ng/uL; PME: 64.2 ng/uL; PAL: 62.9 ng/uL; PDK: 35.7 ng/uL; PDK(2): 81.3 ng/uL; PDK(3): 21.5 ng/uL
07-15-2010 [ top ]
Today, we did minipreps of intron and backbone. Would have also done minipreps of intron plus allergen part but none of those colonies grew.
We also miniprepped Betv1.2 Sense, Betv1.1 Sense, Bet 1.2 Antisense, and Ger Sense of the plates that we grew on Monday. The rest of the allergen panel did not grow properly. What colonies that WERE on the plate grew probably because AMP was not spread properly on the plate. We deduced this because those colonies could not grow in liquid media. We are waiting on more LB amp plates so that we can redo ligations, which is probably the reason for our lack of colonies.
We will do diagnostic digests of all miniprepped DNA.
Results
- Minipreps of PDK+V0120 (LTPS+ PDK and Ger_S + PDK did not grow)
- Made glycerol stocks 500uL of 80% glycerol + 500 uL cells
- Concentrations of pPDK: 414.2 ng/uL; 478.8 ng/uL; 510.2 ng/uL; 119.2 ng/uL
- Tomorrow, sending PDK+V0120 for sequencing
- Concentrations of pPDK: 414.2 ng/uL; 478.8 ng/uL; 510.2 ng/uL; 119.2 ng/uL
- Looking at sequencing results for our other amiRNA parts
- Only GFP worked from our first sequencing order. This time, none of the artificial parts had the right sequence
- Repicking amiRNA colonies to miniprep (6 of each LTP, LTP 0.5, and Bet. We have correct GFP from our first sequencing order)
- Minipreps of Bet 1S (2 colonies); Bet 1.2 S (1 colony); Bet 1.2 A (1 colony); Ger S (3 colonies)
- Redoing Ligations & Transformations of Sense/Antisense Parts once we have LB + amp plates
- Bet 1.1 S: 161 ng/uL & 199.2 ng/uL; Bet 1.2 A: 199.4 ng/uL; Bet 1.2S: 162.5 ng/uL
- Concentrations of GerS: 347.7 ng/uL; 428.6 ng/uL 541.6 ng/uL
- Diagnostic Digest of (PDK+V0120; GERS; BetS; Bet 1.2S/A)
| pPDK (1-4) | pGERSPDK (1-3) | Bet1S (2); Bet2S/A | ||
|---|---|---|---|---|
| DNA | 1 | 1 | 1 | |
| FD Buffer (10x) | 2 | 2 | 2 | |
| diH2O | 15 | 15 | 15 | |
| Xba1 | 1 | 1 | 1 | |
| Pst | 1 | 1 | 1 |
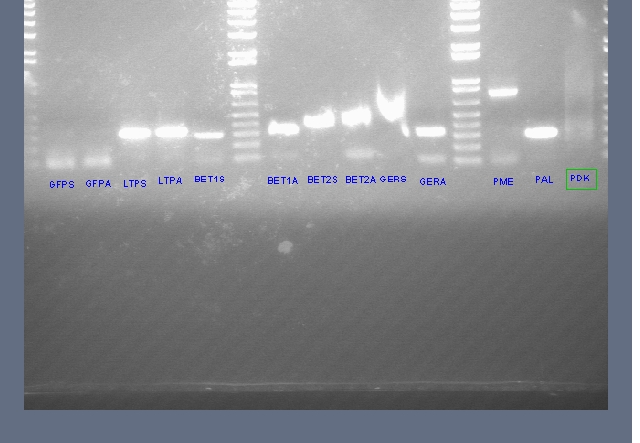
- Ran on gel (Ladder, pdk, V0120 Backbone, Gers (1-3), Bet 1S (1,2), PDK (1-4), Bet 1.2 S, Bet 1.2 A
To send for sequencing: ger s, Bet 1S, PDK 2
- Looking at PCR results for Allergen Panel from Genomic DNA that we extracted (did not work)
Ladder, GFPS, GFPA (These first two should not work), LTPS, LTPA, BetS, BetA, Bet2S, Bet2A, GerS, GerA, PME, PAL, PDK
07-16-2010 [ top ]
More arabadopsis DNA arrived today. We are going to obtain more hpRNA parts and
- Did a second PCR/Purification of our allergen panel from Julia's arabidopsis genomic DNA. It worked!
GFP didn't show up (first 2 lanes). The plants for the extraction weren't the GFP variety so this is expected.
- gel purifying PCR products
- Concentrations: LTPS: LTPA: GERS: GERA: BETS: BETA: BET2S: BET2A: PME: PAL:
- Digested Xba1 Pst1
- Sent BetS, PDK2, and GerS (3) for sequencing
- Miniprep amiRNA
Nanodrop of Miniprepped amiRNA Concentrations
| 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|
| LTP | 320.7 | 359.4 | 290.2 | 238.5 | N/A | 212.4 |
| Bet | 156.8 | 267.7 | 340.0 | 287.4 | 278.9 | 273.5 |
| LTP 0.5 | 296.4 | 268.8 | 244.2 | 212.0 | 292.6 | 208.3 |
Diagnostic Gel: Ladder, Bet1-6, LTP1-LTP4, Ladder; Ladder, LTP6, LTP.5 (1-6), Ladder
07-19-2010 [ top ]
- Got back sequencing results of GerS, BetS, PDK
- All three samples of GerS and PDK sent in matched their corresponding sequences
- Ran PCR of Friday's digested B21. The gel suggested a partial digest: we saw two bands of backbone where we should have seen only one:
- Sending 6 samples of amiRNA (LTP.5), 5 samples of amiRNA (LTP), 6 samples of amiRNA (Bet)
- Second digest of B21 appears to have been more successful:
| V0120 | ||
|---|---|---|
| DNA | ~2.1 ugrams | |
| FD Buffer (10x) | 2 | |
| diH2O | 20-rest | |
| Xba1 | 1 | |
| EcoRI | 1 |
- phosphatized backbone afterwards for 20 min after letting digestion run for an hour
- concentrations
- backbone: (5 copies each ~ 35 ng/uL)
- Bet S/A: ; Bet2S/A: ; GerS/A: ; LTPS/A: ; PME: ; Pal:
| BetS/A | Bet2S/A | LTPS/A | GerS/GerA | Pme | Pal | ||
|---|---|---|---|---|---|---|---|
| diH2O | 10-rest | 10-rest | 10-rest | 10-rest | 10-rest | 10-rest | |
| T4 DNA ligase buffer (10x) | 1 | 1 | 1 | 1 | 1 | 1 | |
| DNA Insert | 3 times excess | 3 times excess | 3 times excess | 3 times excess | 3 times excess | 3 times excess | |
| DNA Backbone | ~50ng | ~50ng | ~50ng | ~50ng | ~50ng | ~50ng | |
| T4 DNA ligase | 1 | 1 | 1 | 1 | 1 | 1 |
- Ligated and transformed
- allergen parts
- Pme/pal
07-20-2010 [ top ]
Today, we plated the transformations that we did yesterday. We are also expecting sequences for amiRNA that we ordered yesterday.
Procedures
- Plating transformants
- Check sequences
Results
Plating Transformations One plate of each allergen S and A and the introns PAL and PME. All but BetS and PME had colonies
Sequencing Results for amiRNA
Not one matched the sequences that we wanted. The customization sequences that we wanted (our miRNA sequence+ a little bit of RS300) were not present in all of the LTP and Bet sequences sent in. Even the Biobrick ends were not present in the sequences except for GFP, from our earlier sequencing order.
What could have gone wrong? We suspect that the RS300 vector that was given to use already contains an interference sequence. The sequence that we used to design our primers are based on an RS300 vector without any inserted interference sequence. Christina has already sent an email to the lab that gave us the RS300 asking if the RS300 contains any interference, possibly GFP. That would explain our results.
It is possible that we will have to go back and redo PCR ~ transformation. These results are all still from the first time that we tried making amiRNA parts, just different colonies.
"c1" and "c2" are the customization (mutagenesis) sites corresponding to the capitalized portions of the primers suggested by the WMD3 tool. "c1" corresponds to the capitalized region of what WMD3 calls primer3 and to the reverse compliment of the capitalized region of primer4. Similarly, "c2" corresponds to the capitalized region of primer1 and the reverse complement of the capitalized region of primer2. For instance, WMD3 gives us the following when asked to design primers for the BET sequence: primer1: gaTTGGTTATATATATCGCACCTtctctcttttgtattcc primer2: gaAGGTGCGATATATATAACCAAtcaaagagaatcaatga primer3: gaAGATGCGATATATTTAACCATtcacaggtcgtgatatg primer4: gaATGGTTAAATATATCGCATCTtctacatatatattcct In the sequence itself, the File c1 Seen c2 Seen ----- First Sequencing Batch ------------ BETF-Fwd.seq TTGGACTGAAGGGAGCTCCC GAGAGCTTCCTTGAGTCCAT BETR-Rev.seq TTGGACTGAAGGGAGCTCCC GAGAGCTTCCTTGAGTCCAT ----- Second Sequencing Batch ----------- bet_2_f-forw TTGGACTGAAGGGAGCTCCC GAGAGCTTCCTTGAGTCCAT bet_3_f-forw TTGGACTGAAGGGAGCTCCC GAGAGCTTCCTTGAGTCCAT ----- Third Sequencing Batch ------------ BET1F-f.seq TTGGACTGAAGGGAGCTCCC GAGAGCTTCCTTGAGTCCAT BET1R-rev.seq TTGGACTGAAGGGAGCTCCC GAGAGCTTCCTTGAGTCCAT BET2F-f.seq TTGGACTGAAGGGAGCTCCC GAGAGCTTCCTTGAGTCCAT BET2R-rev.seq TTGGACTGAAGGGAGCTCCC GAGAGCTTCCTTGAGTCCAT BET3F-f.seq TTGGACTGAAGGGAGCTCCC GAGAGCTTCCTTGAGTCCAT BET3R-rev.seq TTGGACTGAAGGGAGCTCCC GAGAGCTTCCTTGAGTCCAT BET4F-f.seq TTGGACTGAAGGGAGCTCCC GAGAGCTTCCTTGAGTCCAT BET4R-rev.seq TTGGACTGAAGGGAGCTCCC GAGAGCTTCCTTGAGTCCAT BET5F-f.seq TTGGACTGAAGGGAGCTCCC GAGAGCTTCCTTGAGTCCAT BET5R-rev.seq TTGGACTGAAGGGAGCTCCC GAGAGCTTCCTTGAGTCCAT BET6F-f.seq TTGGACTGAAGGGAGCTCCC GAGAGCTTCCTTGAGTCCAT BET6R-rev.seq TTGGACTGAAGGGAGCTCCC GAGAGCTTCCTTGAGTCCAT LTP1F-fwd.seq TTGGACTGAAGGGAGCTCCC GAGAGCTTCCTTGAGTCCAT ... ----- Theoretical Expectations ---------- RS300 Plasmid TTGGACTGAAGGGAGCTCCC GAGAGCTTCCTTGAGTCCAT Bet.ape AGATGCGATATATTTAACCAT TTGGTTATATATATCGCACCT GFP.ape GTACAGCTCGCCGTCCACTAT TTAGTGGTCGGCGAGCTGCAC LTP.ape GTAGTCCTATACAAAGTTAGT TCTAACTATGTATAGGACCAC RS300.ape TATGGACTGAAGGGGACCCGT ACAGGTCCCCTTCTGTCCATT RS300 Backbone TATGGACTGAAGGGGACCCGT ACAGGTCCCCTTCTGTCCATT
07-21-2010 [ top ]
Yesterday, we plated transformants for the allergen panel and introns. Today, we will purify the plasmids from the transformants and send any successes in to Genewiz for sequencing.
Procedures
- Culture Colonies
- Miniprep of colonies
- Diagnostic Digest to check for inserts
- Send working colonies for sequencing
Results
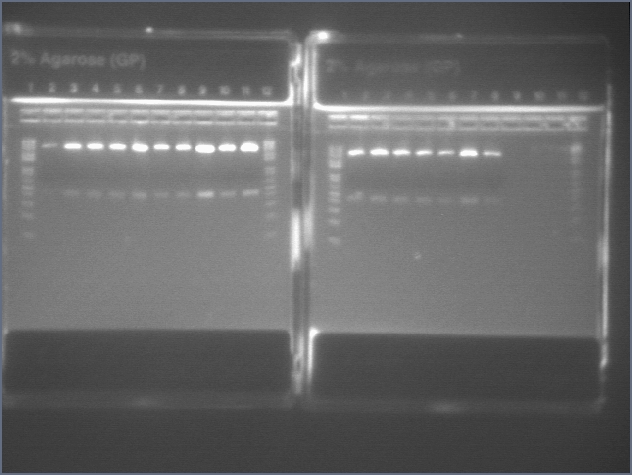
Diagnostic Digest
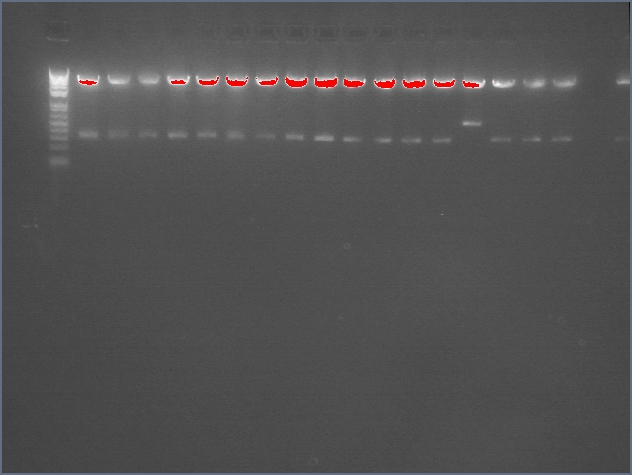
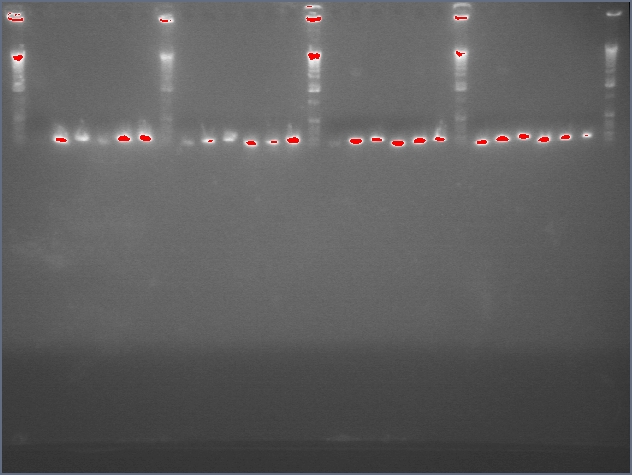
lader; ladder; GerS (1-3); Ger A (1-3); Bet2S (1-3); Bet2A (1-3); BetA (!-3); LTPS (1-3); LTPA (1-3); Pal (1-3); ladder; ladder
(More contrasted image below):
07-22-2010 [ top ]
Today, we are expecting our sequencing results from Genewiz. Once those arrive, we will check the sequences for both the primers and the correct allergen sequence (around 300bps). If the sequences are correct, we will go back to our plasmids and cut the intron out of the plasmid and digest the plasmids containing the sense strands with S, P.
Procedures
- Check Sequencing Results
- Digest Intron with X, P
- Digest Plasmids with Sense with S,P
- Gel Electrophoresis of Digested Plasmids (both Intron and Sense)
- Gel Extraction and Purification of Intron and Antisense
- PCR purification of Sense + Bbone
Results
Sequencing Results
- Ger S (all 3 samples had correct sequence)
- Ger A (all 2 samples had correct sequence)
- Bet 2S (#3 did not work. #2 had correct sequence)
- Bet 2A (all 3 samples had correct sequence)
- LTP S (all 3 samples had correct sequence)
- LTP A (all 2 samples had correct sequence)
- Pal (#3 did not contain plasmids. #1 and #2 had correct sequence)
- PME and Bet v1.1 S did not grow up from the plates, so they were not included in sequencing.
Digest Intron with X, P
Fast Fermentas protocol was used.
Digest Plasmids with S, P Fast Fermentas protocol was used.
Gel electrophoresis of Digested Plasmids We PCR purified the products of the digestion (unnecessarily), losing some DNA. Elution was done with 50μL of EB. Tomorrow, we will run the purified PCR products on a gel and Gel Extract and Purify.
07-23-2010 [ top ]
Today, we will create our very first biobrick vector with both the sense and intron parts. We will use parts digested yesterday, purify them, and ligate them together.
Procedures
- Gel Electrophoresis of Intron and Antisense
- Gel purification of Intron and Antisense inserts
- Ligation of Intron insert and Sense + V0120
- Transform and plate
Results
Gel Electrophoresis
All the Antisense and intron lanes showed, though the last lane (PAL) was faint.
Gel Purification Nanodrop of gel purification had concentrations of:
- LTP: 1.8, 2.9
- Bet v1.1: 6.0, 3.2
- Bet v1.2: 1.8, 0.5, 1.3
- Ger 3: 0.8, -0.5
- Pal: 6.7, 0.7
There is some cause for concern for the lower concentrations, since these small numbers are outside of the range of accuracy.
Ligation of intron insert into Sense + V0120 PME was ligated into all backbones containing sense parts of the allergen panel (8 rxns), followed by PAL ligation into the same backbones. We plated two of each PAL ligation in case that there might be some increased chances of finding growth on plates if more than one plate is done for each ligation. We will check for colonies Saturday.
07-26-2010 [ top ]
Procedures
Last week, we finished ligation together Sense and Intron parts of the hpRNA and transformed them into E. coli. This week, we will extract the plasmids, add in the antisense part, and transform.
Procedures today...
- Culture colonies (done over the weekend)
- Miniprep of V0120 sense + PDK, PAL intron parts
- Diagnostic Digest
We also need to check which parts of the sequence on the team page corresponds to the sense/antisense parts of the gene. There may have been mistakes in the past.
Results
Culture We cultured 38 colonies. All grew but one LTP+PAL and Ger+Pal grew. We labeled the cultures 1- 38 for simplicity sake.
Miniprep
Here are the concentrations of the miniprepped plasmid.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| LTP+PDK | 233.9 | 507.6 | 498.7 | 510.0 | 485.1 | 588.3 | ||||
| LTP+PAL | 471.8 | 586.7 | 500.2 | 449.4 | 492.7 | 430.0 | 521.8 | 605.8 | 499.1 | 299.9 |
| Bet v1+PDK | 430.1 | 519.3 | ||||||||
| Bet v1+PAL | 432.3 | 300.6 | 458.7 | 213.8 | ||||||
| Bet v2+PDK | 270.3 | 223.3 | ||||||||
| Bet v2+PAL | 270.3 | 223.3 | ||||||||
| Bet v2+PDK | 270.3 | 223.3 | ||||||||
| Ger3+PDK | 440.4 | 205.2 | 342.7 | 290.2 | ||||||
| Ger3+PAL | 335.4 | 297.8 | 474.3 | 396.1 | 354.7 | 366.2 |
Diagnostic Digest
We digested the vectors with Spe and Pst1 and ligate with antisense parts (already digested and gel purified). We ran them on the E Gel too long so the lanes do not show anything conclusive, other than that there is digested DNA in the bottom half.
Sequence review clarified which parts of the sequences on the team page corresponded to sense/antisense.
07-27-2010 [ top ]
Procedures
Yesterday, we tried to run a diagnostic digest of the Sense + Intron hpRNA. The gel did not work properly, so we are redoing the gel today. We'll also go ahead and ligate antisense parts into all 38 minipreps that we HOPE contains correct Sense + Intron. Once the gel finishes running, we can go back and dispose of the ligations today that do not contain the correct S+I parts.
Today, we are also going to redo the ligation of Sense + Intron for PDK colonies because the gel that we are redoing today suggests that no PDK worked from last week's ligation.
Procedures
- Diagnostic Digest
- Diagnostic Gel
- Diagnostic Gel of PDK + Sense
- Ligation of Antisense Part into 38 minipreps
Redoing ligations for Sense + PDK
- Digest of pLTP, pBet v1, pGer with S, P
- PCR purification
- Ligation of Sense+PDK
- Transform and Plate
Results
Diagnostic Digest
38 digests of the sense+intron miniprepped plasmid DNA. The digest was done with Slow Digest Eco and Pst
Diagnostic Gel
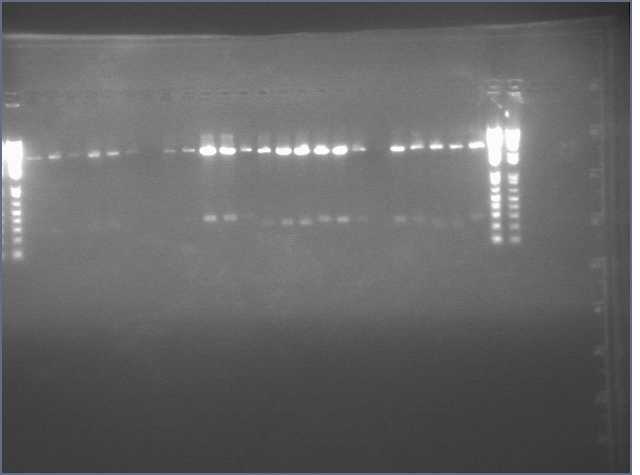
From the digest, we can see that two LTPS+Pal worked (9,11), 3 Ger3S+Pal worked (36-38), 4 Bet2S+Pal Worked (25-28)
Lanes are: 1-6: LTP + PDK (3200 & 1200);
7-16: LTP + PAL (3200 & 540);
17,18: Bet1 + PDK (3200 & 1200);
19-22: Bet1 + PAL (3200 & 540);
23,24: Bet2+PDK (3200 & 1200);
25-28: Bet2 + PAL (3200 & 540);
29-32: Ger3 + PDK (3200 & 1200);
33-38: Ger3 + PAL (3200 & 540);
Image: Ladder (every 6)
Rerun of the lanes that were ligations of PDK + sense because they did not show on the gel above. We feel that there might be a chance that if the Sense+PDK assembly did not work, the 300bp evidence ran off the first gel. This second gel shows that most of the PDK+sense did NOT work. Only three worked -- minipreps 16, 30, and 31.
Ligated with antisense parts and transformed
Digested antisense parts with X, P and pSense+Intron with S, P. Digestion ran for 2 hours, with an additional 20 minutes to de-phosphatize the backbone. Ligations were done for colonies that showed correct Sense+Intron ligations.
| 1-39 | |
|---|---|
| DNA | ~1.5 ug |
| 10x BSA | 3 |
| diH2O | Fill to 30 |
| Spe | 1.5 |
| Pst1 | 1.5 |
| Neb Buffer 2 (10x) | 3 |
Transform and Plate
The standard Turbo Cell transformation protocol was followed.
Redo Ligations for PDK+ Sense
Since the gel showed that most of them did not work.
Digestion of pLTP, pBet v1, and pGer 3 with S, P to prepare them as a back bone. Phosphatase was used. Did not digest pBet v2 because there was not enough.
PCR purify the digested backbones containing Sense.
Nanodrop
pLTP - 24.0 pBet - 17.2 pGer - 42.8
Ligate predigested PDK digested with X, P and ligate into purified backbone. Transform and Plate (6? plates total)
07-28-2010 [ top ]
Procedures
Yesterday, we plated E. coli containing complete hpRNA for Ger, Bet v2, and LTP. Bet v1 did not ligate properly in the earlier Sense+Intron ligation.
Today, we will extract the plasmids from the E. coli and prepare them for sequencing tomorrow. We will also do ligations with the false negatives (Sense + PDK ligate with Antisense).
- Culture colonies
- Miniprep
- Nanodrop
For our false negatives
- Ligate
- Transform
Results
Nanodrop
The format is: Miniprep # , Name of Allergen, concentration in ng/μL of colony 1, concentration in ng/μL of colony 2. All these working ligations are with the PAL intron
- 9, LTP, 97.9, 94.1
- 11, LTP, 77.9, 94.8
- 25, Bet v2, 67.8, 97.8 (there was a mixup with colony 1 and LTP+PDK, but it is now fixed in our electronic notebooks)
- 28, Bet v2, 73.2, 83.6
- 36, Ger3, 76.1, 83.4
- 37, Ger3, 70.0, 75.3
- 38, Ger3, 90.4, 108.3
Transformed more sense+pdk parts for our false negatives (16,30,31)
The parts already have been digested and purified, so today, all we had to do was ligate and transform.
07-29-2010 [ top ]
- Ran a gel for our second attempt at amiRNA stiching (stage 1):
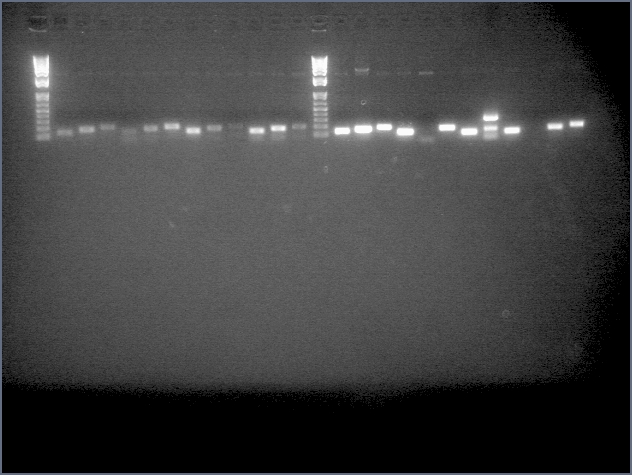 . It seems that the backbone is still present at high concentrations after PCR purification. Maybe this is why last time we had a high incidence of background? This time we're gel purifying.
. It seems that the backbone is still present at high concentrations after PCR purification. Maybe this is why last time we had a high incidence of background? This time we're gel purifying.
- Dignostic Digest
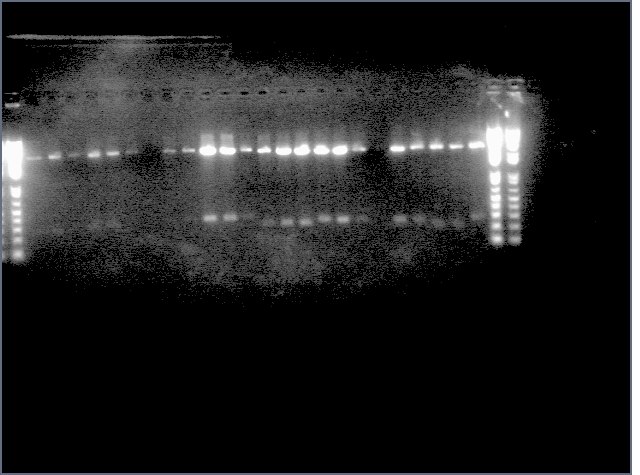
Gel: ladder, 9, 9, 11, 11, 25, 25, 28, 28, ladder, 36, 36, 37, 37, 38, 38, ladder, LTPS+ PDK, LTPS+PDK, Bet2S+PDK, Bet2S+PDK, GerS+PDK, GerS+PDK, GerS+PDK, GerS+PDK
Expected band lengths:
Complete Parts
9,11 (LTPS+Pal+LTPA): 840bp and 3200 bp
25,28 (Bet2S+Pal+Bet2A): 840bp and 3200 bp
36,37,38 (GerS+Pal+GerA): 840bp and 3200 bp
Sense + Intron Parts
LTPS+ PDK: 1200 and 3200 bp
Bet2S+ PDK: 1200 and 3200 bp
GerS+PDK: 1200 and 3200 bp
(9 c1, 25 c2, 36 c1 have a 540 and 3200 band as opposed to a 840 and 3200 band--prbly b/c sense+intron is there but antisense was not ligated in). All intron+sense parts except for LTPS+PDK appear to have 300 bands as well indicating that PDK was not ligated in.
(a small mix up with two of the lids was made and its possible that LTPS+PDK (w/ an 840 and 3200 band is actually the complete Bet2 part so we're sending this in for sequening anyway).
07-30-2010 [ top ]
- Sequencing Results:
LTP ihpRNA parts that worked: 9c2, 11c1, 11c2
Bet 2 ihpRNA parts that worked: 25c1,c2, 28c1,(LTPS+PDK is actually Bet 2 hpRNA)
Ger ihpRNA parts that worked: 36c1,c2, 37c1,2, 38c1,c2
08-03-2010 [ top ]
- Grew up cultures of completed ihpRNA constructs (Bet, LTP, Ger) in pORE expression vector
- amiRNA PCR
- Will look at results of PCR tommorrow
08-04-2010 [ top ]
Tasks
- amiRNA PCR appears to have worked at every Tm we tried:
- Digested V9/V10 to insert our constructs into
- Realized that we hadn't gel purified our ihpRNA inserts
- Gel purification of inserts (entire ihpRNA parts)
Ladder, 9, 11c1, 11c2, ladder, 25c1, 28c1, 28c2, 36c1, 36c2, ladder
Results
- Successfully gel purified our inserts and digested backbones that we will ligate into
08-05-2010 [ top ]
- Gel extracted V9/V10 backbone
Lanes: Ladder, V9, Ladder, V10
Concentrations: V9 (9.4 ng/uL; V10 (16.4 ng/uL)
- Ligated ihpRNA inserts into V9/V10 and transformed
- For our ligations we only used ~ 2uL of backbone (around 18 and 32 ng of backbone)and used a 3x excess of insert
- Verified that amiRNA stitching of Bet, LTP yielded the proper insert with a low level of background through PCR:
Lanes: 1-7: Bet, corresponding to 65.55 degrees C for stitching Tm (even spacing)
Lanes: 8-10: LTP, corresponding to 65.55 degrees C during stitching annealing, even spacing
- Digested
- Bet,LTP inserts with X+P; B21 with X+P+phosphatase
- Ligated, Transformed
08-06-2010 [ top ]
amiRNA
- amiRNA-transformed turbo cells didn't grow in YEB medium. There should be more LB+Amp available by the end of the day. In the meantime, we're digesting some more backbone (8x20ul X+P+B21)
- ligations of Bet/LTP into V0120
ihpRNA
- no colonies on plates from transformations done yesterday after 16 hours (we let these plates grow for longer)
- redoing ligations of ihpRNA inserts in V9/V10
- Redigested and gel purified some backbone
- Concentrations: V9 (5 ng/uL); V10 (6 ng/uL)
- 12 ligations each for V9 and V10
08-09-2010 [ top ]
- DTLs of amiRNA insert were unsuccessful. Yesterday we concluded this was because turbo cells + amp resistance don't grow on YEB media (we were out of LB+amp media). Today we must adopt a new hypothesis as 0/16 colonies grew on LB+amp media. Working theory is that the backbone digestions pictured below have the digested backbone and inserts as the two bands (top to bottom) instead of the undigested & digested backbone (in hindsight, I was trying to ligate into YFP*2, the original insert of B21). The confusion arose from the 1kb ladder, which at first appeared to match another ladder far better than 1kb (see Thursday's entry or thereabouts). This, combined with the fact that the 1kb ladder was hand-labeled, led me to suspect that it was in fact the other ladder (pR322 as pictured [http://openwetware.org/wiki/Image:Ladders.jpg here]). This, in turn, supported the (theorized incorrect) conclusion that the top & bottom bands [http://openwetware.org/wiki/Image:Bbcut1.jpg here] and [http://openwetware.org/wiki/Image:Bbcut2.jpg here] were undigested & digested backbone, respectively, and that the insert had diffused. I've DTL'd again, this time using the upper band (which didn't separate into digested & undigested bands after running for 30m on a .7% agarose gel) for ligation. We'll see on Monday how it went.
- New Plan of action:
- Redigest V9/V10 (three digests of each backbone to use more DNA in ligations)
- Grew up V9/V10+ ihpRNA colonies
- Re pcrd amiRNA and digested to ligate into V9/V10
- Catalog parts to be ready for the registry
08-10-2010 [ top ]
ihpRNA
- Only 2 colonies grew up (V10 9 (37c1) and V10 12 B(38 c2)) ger ihprna constructs
- growing up colonies again in 5 mL cultures w/less antibiotic (50 ng/uL of kan)
- concentrations: V10_9: 21.4 ng/uL V10_12B: 16.9 ng/uL
- Diagnostic Digest (Should see ~ 7 kb band and 840 band)
- Did not see exepcted bands (waiting on cultures grown up today to miniprep and hopefully transform into agrobacterium tommorow)
amiRNA
- Ligated into V9/V10 and transformed
08-11-2010 [ top ]
- Minipreps of (V95, V91, V99, V99b, V1011, V107B, V1012, V109b)
- Concentrations (20-60 ng/uL range)
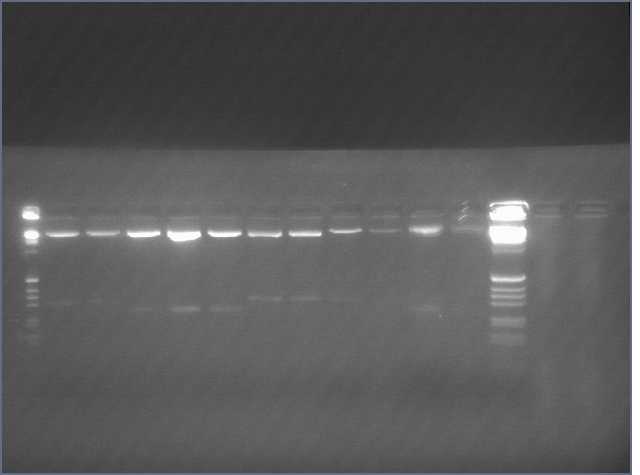
- Diagnostic Digest (should see ~ 7 kb and 840 bp)
Ladder, V9 5 (Bet2), V91(LTP), V99 (Ger), V99B (Ger), V1011 (Ger), V107b(Ger), V1012 (Ger), V109B (Ger)
V10 7B (Ger worked)
08-12-2010 [ top ]
ihpRNA
- Minipreps of more V9/V10 colonies
- Diagnostic Digest of minipreps w/ Eco and Pst (Should see ~840 and 7000 bp)
ladder, V92, V92b, V94, V94b, V95, V96b, V97, V97b, V98, V98b
Lanes 2&3: LTP (11c1); Lanes 4&5: Bet2 (25c1); Lane 6: Bet2 (25c2); Lane 7: Bet2 (28c1); Lanes 8&9: Ger (36c1); Lanes 10&11 Ger (36c2)
- Digested V9/V10 backbone for ligations w/ Eco & Pst (redoing ihpRNA ligations into V9/V10)
amiRNA
- Ligated into V9/V10 and transformed (mutagenesis sites confirmed through pcr using 20 bp sepecific primers)
- still need to be sequenced
08-13-2010 [ top ]
- Organized parts so we could find them when we came back
<Note: remember to add more pics/less text>
 "
"

 [click to enlarge]
[click to enlarge]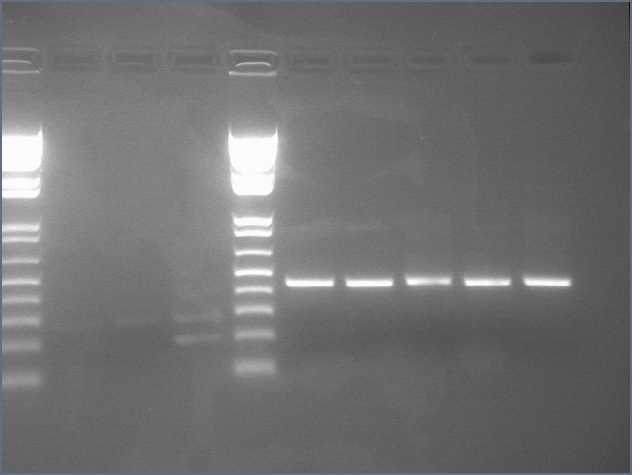

 [click to enlarge]
[click to enlarge]
 [click to enlarge]
[click to enlarge] [click to enlarge]
[click to enlarge] [click to enlarge]
[click to enlarge] [click to enlarge]
[click to enlarge]