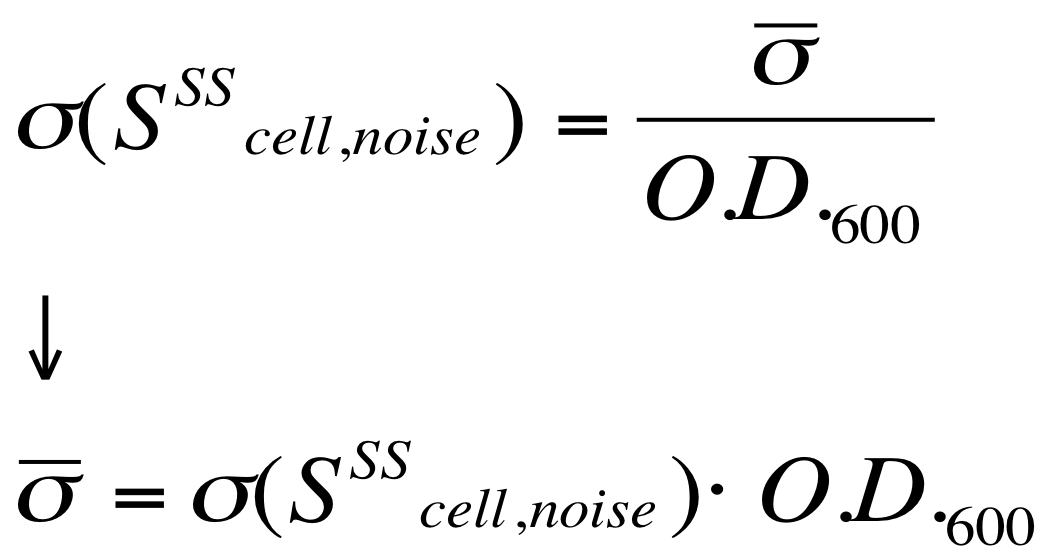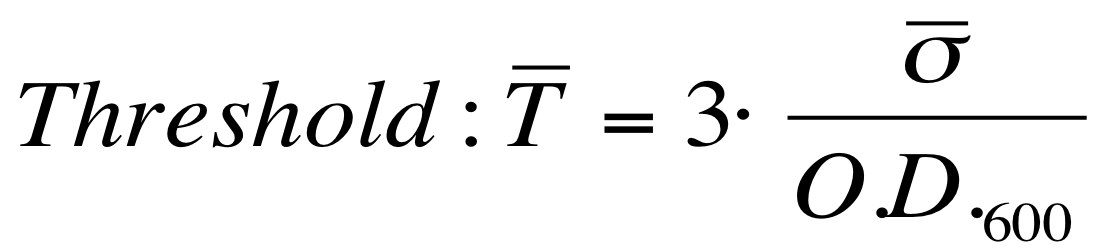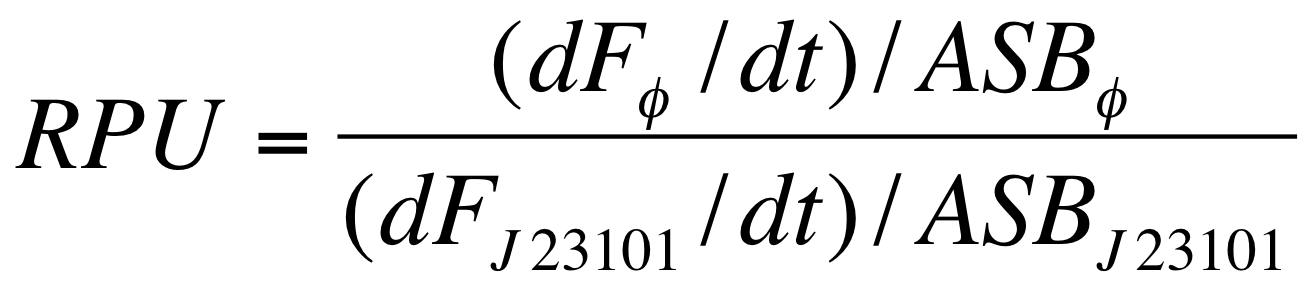|
|
| Line 9: |
Line 9: |
| | <table border="0" align="center" width="100%"><tr><td align="justify" valign="top" style="padding:20px"> | | <table border="0" align="center" width="100%"><tr><td align="justify" valign="top" style="padding:20px"> |
| | <html><p align="center"><font size="4"><b>CHARACTERIZATION</b></font></p></html><hr><br> | | <html><p align="center"><font size="4"><b>CHARACTERIZATION</b></font></p></html><hr><br> |
| - |
| |
| - | =All our parts=
| |
| - |
| |
| - | ===Our new parts===
| |
| - | '''Self-inducible promoters''''
| |
| - |
| |
| - | ===<b>Exploiting quorum sensing mechanism...</b>===
| |
| - |
| |
| - | Different studies demonstrated that bacteria, such as V. fischeri, can communicate through a mechanism called quorum sensing and regulate genic expression relying on cell culture density. One of the most studied organisms is ''V. fischeri'', for which the quorum sensing is regulated by two genes: luxI and luxR. The first one encodes a protein responsible for the synthesis of homoserine-lactone, also called the auto-inducer. The second one encodes a protein capable to bind the HSL, once it has reached a critical concentration, making a complex that activates the transcription of genes under the regulation of the ''lux pR'' promoter. As the concentration of HSL is an increasing function of cell culture density, the induction of the ''lux pR'' promoter occurs only when the O.D.600 reaches a threshold concentration.
| |
| - | Taking inspiration from this natural mechanism, a library of self inducible promoters was built, exploiting the quorum sensing in ''E. coli''.
| |
| - |
| |
| - |
| |
| - | [[Image:pv_SenderReceiverAntenna.png|450px|thumb|center|Figure 1 - Sender/receiver behaviour exploited to obtain self-inducible devices]]
| |
| - |
| |
| - | ===<b>Parts and system overview</b>===
| |
| - |
| |
| - | Two BioBrick parts already present in the registry were used in this module. The luxI coding sequence (<partinfo>BBa_K081008</partinfo>) was assembled upstream of the double terminator <partinfo>BBa_B0015</partinfo>, thus obtaining the fundamental part to build self-inducible circuits, <partinfo>BBa_K300009</partinfo> (Fig. 2).
| |
| - | [[Image:pv_K300009.png|230px|thumb|center|Figure 2 - BBa_K300009]]
| |
| - |
| |
| - | This part was used as signal generator, while the signal receiver part is <partinfo>BBa_F2620</partinfo>. In order to build a library of self-inducible promoters, another foundamental device was obtained by assembling <partinfo>BBa_K300009</partinfo> upstream of <partinfo>BBa_F2620</partinfo>, thus obtaining the part <partinfo>BBa_K300010</partinfo> (Fig. 3).
| |
| - |
| |
| - | [[Image:pv_K300010.png|450px|thumb|center|Figure 3 - BBa_K300010]]
| |
| - |
| |
| - | This systems have the behaviour shown in figure (Fig. 4). LuxR is contitutively produced under the regulation of pTet promoter, while LuxI is produced under the control of a different constitutive promoter. ''lux pR'' is activated when the [HSL] present in the medium is over a threshold, that we evaluated (more details in "Threshold evaluation for ''lux pR'' avtivation" Section).
| |
| - |
| |
| - | [[Image:pv_I80_working.png|450px|thumb|center|Figure 4 - Self-inducible device behaviour]]
| |
| - |
| |
| - |
| |
| - |
| |
| - | ===<b>Regulation of signal protein productuion</b>===
| |
| - |
| |
| - | This part was assembled downstream of different constitutive promoters, thus obtaining a signal molecule generator. The choice of constitutive promoters was performed between the ones belonging to the [http://partsregistry.org/Part:BBa_J23101 Anderson’s promoters collection] ; we chose promoters according to their activities reported in the Registry of Standard Biological Parts, in order to have a thick mesh:
| |
| - |
| |
| - |
| |
| - | {| align='center' border='1'
| |
| - | |<b>Promoter</b> || <b>Strength (a.u.)<br>reported in the Registry</b>
| |
| - | |-
| |
| - | |<partinfo>BBa_J23100</partinfo> || 2547
| |
| - | |-
| |
| - | |<partinfo>BBa_J23101</partinfo>||1791
| |
| - | |-
| |
| - | |<partinfo>BBa_J23105</partinfo>||623
| |
| - | |-
| |
| - | |<partinfo>BBa_J23106</partinfo>||1185
| |
| - | |-
| |
| - | |<partinfo>BBa_J23110</partinfo>||844
| |
| - | |-
| |
| - | |<partinfo>BBa_J23114</partinfo>||256
| |
| - | |-
| |
| - | |<partinfo>BBa_J23116</partinfo>||396
| |
| - | |-
| |
| - | |<partinfo>BBa_J23118</partinfo>||1429
| |
| - | |}
| |
| - |
| |
| - | Before constructing the signal generators, <partinfo>BBa_K300009</partinfo> under the regulation of one of these constitutive promoters, we evaluated their activities in Relative Promoter Units (R.P.U.) according to [[Team:UNIPV-Pavia/Parts/Characterization#Data analysis for RPU evaluation|Data analysis for RPU evaluation]], using the reporter protein R.F.P. in different experimental conditions (plasmids’ copy number and growth medium) :
| |
| - | *high copy number plasmids and LB;
| |
| - | *high copy number plasmids and M9;
| |
| - | *low copy number plasmids and M9.
| |
| - | It was not possible to evaluate promoters activities in low copy number plasmids and LB because the RFP activity was too weak and not detectable and discernible from the background.
| |
| - | Red Fluorescent Protein (RFP) fluorescence and Optical Density at 600nm (O.D.600) were measured by Tecan Infinite F200 as reported in [[Team:UNIPV-Pavia/Parts/Characterization#Microplate reader experiments for constitutive promoters (R.P.U. evaluation) - Protocol #2|Microplate reader experiments for constitutive promoters (R.P.U. evaluation) - Protocol #2]] and data were analyzed as reported in DATA Analysis RPU; results are shown here:
| |
| - |
| |
| - | {| align='center'
| |
| - | |[[Image:pv_RPU_HC_LB.png|330px|thumb|center|Figure 5 - R.P.U. of some promoters from Anderson promoters' collection, LB medium and high copy plasmid (<partinfo>BBa_J61002</partinfo>) ]]||[[Image:pv_RPU_HC_M9.png|330px|thumb|center|Figure 6 - R.P.U. of some promoters from Anderson promoters' collection, M9 medium and high copy plasmid (<partinfo>BBa_J61002</partinfo>)]]
| |
| - | |}
| |
| - | {| align='center'
| |
| - | |[[Image:pv_RPU_LC_M9.png|330px|thumb|center|Figure 7 - R.P.U. of some promoters from Anderson promoters' collection, LB medium and high copy plasmid (<partinfo>pSB4C5</partinfo>)]]
| |
| - | |}
| |
| - |
| |
| - | We observed that the Registry ranking is not always respected. As an example, <partinfo>BBa_J23110</partinfo> in high copy plasmid is stronger than <partinfo>BBa_J23118</partinfo>, in contrast with the ranking of the Registry.
| |
| - |
| |
| - | After the evaluation of promoters’ activity, signal generators were constructed in high copy and low copy plasmids: <partinfo>BBa_K300009</partinfo> and <partinfo>BBa_K300010</partinfo> were assembled downstream the above mentioned promoters, thus obtaining the following parts:
| |
| - |
| |
| - | {| border='1' align='center'
| |
| - | | '''BioBrick''' ||'''Description'''
| |
| - | |-
| |
| - | | <partinfo>BBa_K300030</partinfo>|| [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23118
| |
| - | |-
| |
| - | | <partinfo>BBa_K300028</partinfo>|| [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23110
| |
| - | |-
| |
| - | | <partinfo>BBa_K300029</partinfo>|| [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23116
| |
| - | |-
| |
| - | | <partinfo>BBa_K300025</partinfo>|| [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23101
| |
| - | |-
| |
| - | | <partinfo>BBa_K300026</partinfo>|| [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23105
| |
| - | |-
| |
| - | | <partinfo>BBa_K300027</partinfo>|| [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23106
| |
| - | |-
| |
| - | | <partinfo>BBa_K300017</partinfo>|| [[Image:pv_SignalGeneratorSensorDevice.png|300px]]<br>J23118
| |
| - | |-
| |
| - | | <partinfo>BBa_K300014</partinfo>|| [[Image:pv_SignalGeneratorSensorDevice.png|300px]]<br>J23110
| |
| - | |-
| |
| - | | <partinfo>BBa_K300015</partinfo>|| [[Image:pv_SignalGeneratorSensorDevice.png|300px]]<br>J23114
| |
| - | |-
| |
| - | | <partinfo>BBa_K300016</partinfo>|| [[Image:pv_SignalGeneratorSensorDevice.png|300px]]<br>J23116
| |
| - | |-
| |
| - | | <partinfo>BBa_K300012</partinfo>|| [[Image:pv_SignalGeneratorSensorDevice.png|300px]]<br>J23105
| |
| - | |}
| |
| - |
| |
| - |
| |
| - | Some of the promoters probably induce a production of LuxI that is injurious for ''E. coli'', so it wasn’t possible to exploit all the combinations.
| |
| - | For every part, a measurement system was built, exploiting the production of the reporter gene GFP to evaluate the trascription initiation point for every promoter. Many different combination were explored, in order to provide a library of promoters capable of initiate transcription at the desired culture density.
| |
| - |
| |
| - | ===<b>Quantification of the HSL produced</b>===
| |
| - |
| |
| - | The new parts were, thus, characterized, measuring the HSL concentration after a 6 hour growth of the cultures. All the details are available in [[Team:UNIPV-Pavia/Parts/Characterization#Microplate reader experiments for 3OC6-HSL quantification by means of BBa_T9002 sensor - Protocol #3|this section]]
| |
| - | <partinfo>BBa_T9002</partinfo> was contained in <partinfo>pSB1A2</partinfo> plasmid experiments were performed on ''E. coli'' TOP10.
| |
| - |
| |
| - | The amount of 3OC6-HSL produced after 6 hours growth by the parts contained in high copy plasmid <partinfo>pSB1A2</partinfo> is reported in Figure 8 and in the table:
| |
| - |
| |
| - | {| align='center'
| |
| - | |[[Image:pv_HCT9002sensor.png|500px|thumb|center|Figure 8 - <partinfo>BBa_T9002</partinfo> calibration curve for detection of [HSL] produced in high copy plasmid]]
| |
| - | |}
| |
| - |
| |
| - | {|
| |
| - | | ''BioBrick'' || ''Wiki name''|| '''E. coli''' ''strain'' || [HSL]
| |
| - | |-
| |
| - | | <partinfo>BBa_K300030</partinfo> || I14|| DH5alpha || 0,7 uM
| |
| - | |-
| |
| - | | <partinfo>BBa_K300028</partinfo> || I15|| DH5alpha || 0,04 uM
| |
| - | |-
| |
| - | | <partinfo>BBa_K300029</partinfo> || I16|| DH5alpha || not detected
| |
| - | |-
| |
| - | | <partinfo>BBa_K300025</partinfo> || I17|| DH5alpha || 0,09 uM
| |
| - | |-
| |
| - | | <partinfo>BBa_K300026</partinfo> || I18|| DH5alpha || not detected
| |
| - | |-
| |
| - | | <partinfo>BBa_K300027</partinfo> || I19|| DH5alpha || 0,002 uM
| |
| - | |}
| |
| - |
| |
| - | The amount of 3OC6-HSL produced after 6 hours growth by the parts contained in low copy plasmid <partinfo>pSB4C5</partinfo> is reported in Figure 9 and in the table:
| |
| - |
| |
| - | {| align='center'
| |
| - | |[[Image:pv_LCT9002sensor.png|500px|thumb|center|Figure 9 - <partinfo>BBa_T9002</partinfo> calibration curve for detection of [HSL] produced in low copy plasmid]]
| |
| - | |}
| |
| - |
| |
| - |
| |
| - | {|
| |
| - | | ''BioBrick'' || ''Wiki name''|| '''E. coli''' ''strain'' || [HSL]
| |
| - | |-
| |
| - | | <partinfo>BBa_K300030</partinfo> || I14|| DH5alpha || 0,005 uM
| |
| - | |-
| |
| - | | <partinfo>BBa_K300028</partinfo> || I15|| DH5alpha || 0,002 uM
| |
| - | |-
| |
| - | | <partinfo>BBa_K300029</partinfo> || I16|| DH5alpha || not detected
| |
| - | |-
| |
| - | | <partinfo>BBa_K300025</partinfo> || I17|| DH5alpha || 0,003 uM
| |
| - | |-
| |
| - | | <partinfo>BBa_K300026</partinfo> || I18|| DH5alpha || not detected
| |
| - | |-
| |
| - | | <partinfo>BBa_K300027</partinfo> || I19|| DH5alpha || not detected
| |
| - | |}
| |
| - |
| |
| - | ===<b>Modulation of plasmid copy number</b>===
| |
| - |
| |
| - | Besides the use of constitutive promoters of different strength to regulate the production of the signal molecule, another parameter took into consideration is the plasmid copy number. Signal generator and sensor device were assembled in an unique part (such as <partinfo>BBa_K300017</partinfo>, <partinfo>BBa_K300014</partinfo>, <partinfo>BBa_K300015</partinfo>, <partinfo>BBa_K300016</partinfo> and <partinfo>BBa_K300012</partinfo>) beared on high copy number plasmid <partinfo>pSB1A2</partinfo> or low copy number plasmid <partinfo>pSb4C5</partinfo>. A third alternative was the assembly of signal generator on a low copy number plasmid (<partinfo>pSB4C5</partinfo>) and the receiver device on high number plasmid (<partinfo>pSB1A2</partinfo>). The evaluated combinations are summurized in Fig. 10, 11 and 12.
| |
| - |
| |
| - | {| align='center'
| |
| - | |[[Image:pv_HCHC.png|330px|thumb|center|Figure 10 - Both sender and receiver are assembled on high copy number plasmid ]]||[[Image:pv_LCLC.png|330px|thumb|center|Figure 11 - Both sender and receiver are assembled on low copy number plasmid]]
| |
| - | |}
| |
| - | {| align='center'
| |
| - | |[[Image:pv_HCLC.png|330px|thumb|center|Figure 12 - Sender part in low copy number plasmid and receiver on high copy number plasmid]]
| |
| - | |}
| |
| - |
| |
| - | The circuits we obtained and tested are summarized here:
| |
| - |
| |
| - |
| |
| - | {| border='1' align='center'
| |
| - | | '''BioBrick'''<br> '''Sender''' ||'''Description ''' || '''Sender Vector''' || '''<partinfo>BBa_F2620</partinfo><br> Receiver vector'''|| '''BioBrick composite part'''
| |
| - | |-
| |
| - | | <partinfo>BBa_K300030</partinfo>
| |
| - | | [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23118
| |
| - | |colspan="2" align='center'| <partinfo>pSB1A2</partinfo><br>HC
| |
| - | |<partinfo>BBa_K300017</partinfo>
| |
| - | |-
| |
| - | | <partinfo>BBa_K300028</partinfo>
| |
| - | | [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23110
| |
| - | |colspan='2' align='center'| <partinfo>pSB1A2</partinfo><br>HC
| |
| - | |<partinfo>BBa_K300014</partinfo>
| |
| - | |-
| |
| - | | <partinfo>BBa_K300029</partinfo>
| |
| - | | [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23116
| |
| - | | colspan='2' align='center'| <partinfo>pSB1A2</partinfo><br>HC
| |
| - | |<partinfo>BBa_K300016</partinfo>
| |
| - | |-
| |
| - | | <partinfo>BBa_K300026</partinfo>
| |
| - | | [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23105
| |
| - | | colspan='2' align='center'| <partinfo>pSB1A2</partinfo><br>HC
| |
| - | |<partinfo>BBa_K300012</partinfo>
| |
| - | |-
| |
| - | | xxx|| [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23114
| |
| - | |colspan='2' align='center'| <partinfo>pSB1A2</partinfo><br>HC
| |
| - | |<partinfo>BBa_K300015</partinfo>
| |
| - | |-
| |
| - | | <partinfo>BBa_K300030</partinfo>
| |
| - | | [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23118
| |
| - | |colspan='2' align='center'| <partinfo>pSB4C5</partinfo><br>LC
| |
| - | |<partinfo>BBa_K300017</partinfo>
| |
| - | |-
| |
| - | | <partinfo>BBa_K300028</partinfo>
| |
| - | | [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23110
| |
| - | | colspan='2' align='center'| <partinfo>pSB4C5</partinfo><br>LC
| |
| - | |<partinfo>BBa_K300014</partinfo>
| |
| - | |-
| |
| - | | <partinfo>BBa_K300029</partinfo>
| |
| - | | [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23116
| |
| - | | colspan='2' align='center'| <partinfo>pSB4C5</partinfo><br>LC
| |
| - | |<partinfo>BBa_K300016</partinfo>
| |
| - | |-
| |
| - | | <partinfo>BBa_K300026</partinfo>
| |
| - | | [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23105
| |
| - | | colspan='2' align='center'| <partinfo>pSB4C5</partinfo><br>LC
| |
| - | |<partinfo>BBa_K300012</partinfo>
| |
| - | |-
| |
| - | | <partinfo>BBa_K300030</partinfo>
| |
| - | | [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23118
| |
| - | |<partinfo>pSB4C5</partinfo><br>LC
| |
| - | | <partinfo>pSB1A2</partinfo><br>HC
| |
| - | | Parts are contained in two different vectors
| |
| - | |-
| |
| - | | <partinfo>BBa_K300028</partinfo>
| |
| - | | [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23110
| |
| - | |<partinfo>pSB4C5</partinfo><br>LC
| |
| - | | <partinfo>pSB1A2</partinfo><br>HC
| |
| - | | Parts are contained in two different vectors
| |
| - | |-
| |
| - | | <partinfo>BBa_K300029</partinfo>
| |
| - | | [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23116
| |
| - | |<partinfo>pSB4C5</partinfo><br>LC
| |
| - | | <partinfo>pSB1A2</partinfo><br>HC
| |
| - | | Parts are contained in two different vectors
| |
| - | |-
| |
| - | | <partinfo>BBa_K300025</partinfo>
| |
| - | | [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23101
| |
| - | |<partinfo>pSB4C5</partinfo><br>LC
| |
| - | | <partinfo>pSB1A2</partinfo><br>HC
| |
| - | | Parts are contained in two different vectors
| |
| - | |-
| |
| - | | <partinfo>BBa_K300026</partinfo>
| |
| - | | [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23105
| |
| - | |<partinfo>pSB4C5</partinfo><br>LC
| |
| - | | <partinfo>pSB1A2</partinfo><br>HC
| |
| - | | Parts are contained in two different vectors
| |
| - | |-
| |
| - | | <partinfo>BBa_K300027</partinfo>
| |
| - | | [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23106
| |
| - | |<partinfo>pSB4C5</partinfo><br>LC
| |
| - | | <partinfo>pSB1A2</partinfo><br>HC
| |
| - | | Parts are contained in two different vectors
| |
| - | |}
| |
| - |
| |
| - | ===<b>Results</b>===
| |
| - |
| |
| - | The following measurement systems were realized assembling GFP downstream of each self-inducible device. The parts characterized are reported in this table:
| |
| - |
| |
| - | {| border='1' align='center'
| |
| - | | '''BioBrick device'''
| |
| - | | '''Measurement system'''
| |
| - | |-
| |
| - | |<partinfo>BBa_K300030</partinfo> in <partinfo>pSb1A2</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23118
| |
| - | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A2</partinfo><br>[[Image:pv_T9002.png|150px]]<br>in <partinfo>pSB1A2</partinfo>
| |
| - | |-
| |
| - | |<partinfo>BBa_K300030</partinfo> in <partinfo>pSb1A2</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23118
| |
| - | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A2</partinfo><br>[[Image:pv_T9002.png|150px]]<br>in <partinfo>pSB1A2</partinfo>
| |
| - | |}
| |
| - |
| |
| - | Cultures of ''E. coli'' TOP10 bearing the plasmids containing the self-inducible devices expressing G.F.P. were grown according to [[Team:UNIPV-Pavia/Parts/Characterization#Microplate reader experiments for self-inducible promoters - Protocol #1|this protocol]] and all data collected were analyzed as explained in [[Team:UNIPV-Pavia/Parts/Characterization#Data analysis for self-inducible promoters (initiation-treshold determination)|this section]]
| |
| - |
| |
| - |
| |
| - |
| |
| - | Dubling times were estimated as explained [[Team:UNIPV-Pavia/Parts/Characterization#Doubling time evaluation|here]]
| |
| - | Thus, these BioBrick parts can be used to express recombinant proteins without adding an inducer to trigger the transcription of their genes; in large-scale production of such proteins this strategy could be also cost saving.
| |
| | | | |
| | =Growth conditions= | | =Growth conditions= |
 "
"