Team:UNIPV-Pavia/Project/solution
From 2010.igem.org
(→Integrative standard vectors for E. coli) |
|||
| Line 66: | Line 66: | ||
<br> | <br> | ||
<table align="center" border="0" width="80%"> | <table align="center" border="0" width="80%"> | ||
| - | + | =Self-inducible promoters= | |
| - | + | =Integrative standard vector for E. coli= | |
The integration of the genetic circuits of interest into the microbial host genome can eliminate the need of expensive selection techniques, such as antibiotics or auxotrophic media, in cell cultures. | The integration of the genetic circuits of interest into the microbial host genome can eliminate the need of expensive selection techniques, such as antibiotics or auxotrophic media, in cell cultures. | ||
| Line 92: | Line 92: | ||
The ''guide'' is the DNA sequence that is used to target the ''passenger'' into a specific locus in the genome. | The ''guide'' is the DNA sequence that is used to target the ''passenger'' into a specific locus in the genome. | ||
| - | + | =Integrative standard vectors for yeast= | |
| - | + | =Self-cleaving affinity tags to easily purify proteins= | |
Revision as of 10:30, 22 October 2010
|
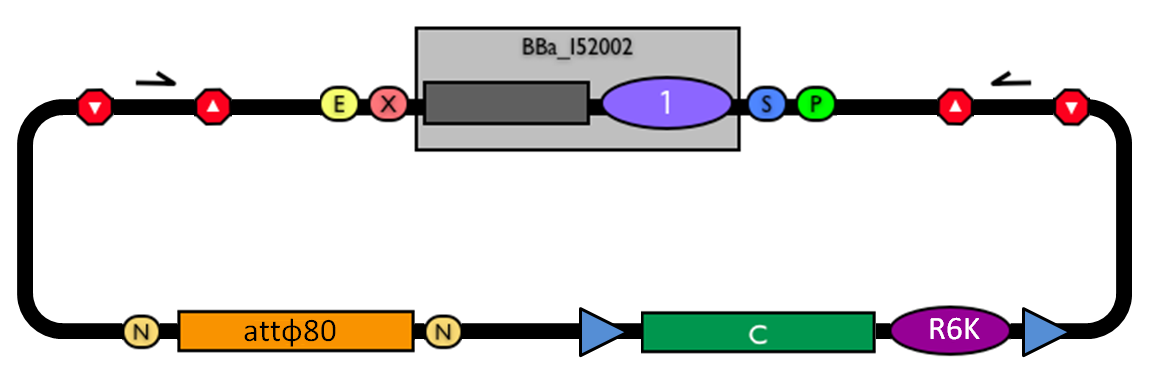
This vector can be considered as a base vector, which can be specialized to target the desired integration site in the host genome. The default version of this backbone has the bacteriophage Phi80 attP (<partinfo>BBa_K300991</partinfo>) as integration site. This vector enables multiple integrations in different positions of the same genome.
GlossaryThe passenger is the desired DNA part to be integrated into the genome. The guide is the DNA sequence that is used to target the passenger into a specific locus in the genome. Integrative standard vectors for yeastSelf-cleaving affinity tags to easily purify proteins
|
 "
"