File:ICLdeticon.png
From 2010.igem.org
(→Candidate Proteins: LytC and CwlC) |
|||
| Line 10: | Line 10: | ||
===Candidate Proteins: LytC and CwlC=== | ===Candidate Proteins: LytC and CwlC=== | ||
| + | |||
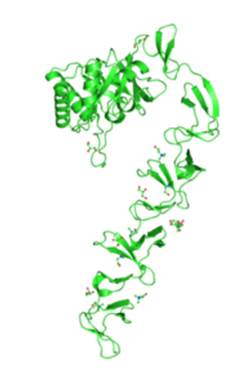
| + | [[Image:Picture1.png|600px|thumb|right|Crystal structure of lytC from EMBA-EBI, 2010]] | ||
*'''What do we have to consider?''' | *'''What do we have to consider?''' | ||
Revision as of 16:16, 19 October 2010
| Detection Module | Signaling Module | Fast Response Module |
| We decided to design a new mechanism for parasite detection - by using the proteases they release. A novel protein bound to the cell surface, with a signaling peptide attached via a protease cleavage site. When the protease comes along, the signal peptide is released, allowing it to activate our signaling module. | To transduce the signal we used a quorum sensing system of a gram positive bacterium. The two component signal transduction system taken from S. pneumoniae transfers our peptide signal into the cell, activating the fast response module. | Our fast response mechanism is based around using two enzymatic amplification steps involving a transcripted enzyme, a deactivated enzyme and a presynthesised substrate. This greatly reduces the time required for producing a recognisable output, enabling useful field testing kits. |
The Detection Module: Based on a Cleavable Surface Protein
In order to set off the fast response module in the presence of the schistosoma cercaria, we designed a protein that carries our signal peptide ComC out of the cell where the protease has access to it. The protease can then proceed to cleave ComC off the protein, allowing quorum sensing via the the ComCDE system to take place. One big problem we have to overcome is the cell wall that will obstruct the protease’s access to the protein carrying the signal peptide. The Detection module consists of a cell wall anchor, a signal peptide called ComC, and a linker that connects the two and is specifically designed to be cleaved by the protease we want to detect.
Candidate Proteins: LytC and CwlC
- What do we have to consider?
There are several restrictions to the choice of surface proteins we have to consider. First of all the use of peptides as quorum sensing signals is mainly restricted to gram positive bacteria such as B. subtilis. Additionally we had to overcome the problem of false positive activation of our system that could be caused when ComC comes in close proximity to the membrane bound receptor ComD. Therefore we decided to choose a cell wall bound protein rather than a membrane bound proteins as originally planned. Finally, because of the nature of the protease and it cleavage specificity we have to attach ComC to the C-terminus of our protein in order to preserve the original ComC sequence after cleavage has occurred.
- What are our candidate genes?
The two most promising proteins we have identified are CwlB and CwlC, both of which found naturally on the cell wall of Bacillus subtilis. CwlB, also known as lytC, is a major autolysin involved in cell wall turn-over and more specifically responsible for breaking down peptidoglycans (N-acetylmuramoyl-L-alanine amidase). Cells producing lytC are uniformly coated with lytC after the middle of the exponential phase. CwlC is also a hydrolase involved in peptidoglycan break down, however it role is less well understood although it too acts as N-acetylmuramoyl-L-alanine amidase playing an important role in mother-cell lysis during sporulation [http://jb.asm.org/cgi/reprint/185/22/6666 Yamamoto et al.]. Both proteins have in common that they use a cell wall binding domain (CWB) consisting of several repeats to bind non-covalently to the bacterial cell wall using charged amino acid residues. The CWBs themselves do not show sequence homology and are of very different sizes, but as they function in a similar manner, both can be eluded from the cell wall by solution with a high osmolarity as described by [http://pubs.acs.org/doi/pdf/10.1021/bi050624n Mishima et al.].
- Why should they work?
Several papers, especially those by Kobayashi et al., have demonstrated that protein domains fused the cell wall binding domains of lytC and cwlC are expressed correctly and localized on the surface of the cell whilst maintaining their catalytic ability.
[http://www3.interscience.wiley.com/journal/119078114/abstract?CRETRY=1&SRETRY=0 Tsuchiya et al. 1999] [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6VSD-426YS50-D&_user=217827&_coverDate=12%2F31%2F2000&_rdoc=1&_fmt=high&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000011279&_version=1&_urlVersion=0&_userid=217827&md5=4764a7bdabc3649925f082f989e835f5 Kobayashi et al. 2000] [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6VSD-46G3T2V-4&_user=217827&_coverDate=01%2F31%2F2002&_rdoc=1&_fmt=high&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000011279&_version=1&_urlVersion=0&_userid=217827&md5=c807c9de3e3579777b08fdd77cd32294 Kobayashi et al. 2002] [http://jb.asm.org/cgi/reprint/185/22/6666 Yamamoto et al. 2003]
Additionally, since we do not intend to knock out the original gene from the B. subtilis strain we will not interfere with it normal expression, avoiding expression at the wrong time or overexpression, and since we aim to remove the catalytic domain from CwlB or CwlC our fusion protein should lose their cell wall turn-over functions and only act as cell wall anchors.
What is the structure of our construct?
We decided to use the CWB of lytC as the basis of our detection module. This decision was based on the fact that more research and recombinant testing of LytC has been done than on CwlC. The gram positive bacteria automatically recognise CWB as such an transport the protein out of the cell where it attaches non-covalently to the cell wall. There is the option to reduce the size of the CWB as it consists of several repeats all but one of which could be removed, however this has not been tested before and thus might not work. Adding the catalytic domain of the protein might help correct localization, but studies have shown that it is not necessary and furthermore ectopic expression of protein at the wrong time might be harmful to the organism. Therefore we have decided to use the cell wall binding domain with the original number of repeats but remove the catalytic domain completely. Next we added a linker to connect the CWB to the signal peptide ComC. The linker has to fulfil various criteria: It has to contain a specific sequence, recognised by the protease we want to detect, in our case cercarial elastase. Furthermore it has to stretch the ComC far enough away from the CWB to allow access of the protease to the recognition sequence. Additionally the linker should not be too flexible as to allow contact of ComC with its receptor ComC as this might set off false positives. This linker could either be a simple glycine repeat or, like Yamamoto et al. suggested, this sequence: SRGSRA (for lytC). However we opted for two linkers suggested by another paper, one helical and one flexible, as well as a pure glycin linker. The third component of the detection module is the signal peptide that has to be linear to allow easy expression and avoid the need for post translational modification. The ComCDE quorum sensing system was the best candidate and well be discussed in more detail in the second module. In order to get a particularly strong signal we considered using several repeats of ComC however we ultimately decided not do so for several reasons: The cercarial elastase cleaves at the C-terminus of its recognition sequence. If several ComC molecules were attached to each other these four amino acids would remain attached to the ComC molecule and potentially interfere with recognition of ComC by its receptor ComD. Additionally the receptor requires a concentration of only 10ng/ml of ComC for detection which, as our models indicate should be reached easily with just one ComC molecule per surface protein.
How do we test for correct expression?
In order to be able to detect out protein we want to use His-tags which will only be added to test constructs but not the final module as it would most likely interfere with the module’s function. The his-tags have been added to the end of ComC, probably inhibiting correct detection of ComC by the two component system comD/E. Using a his-tag allow us to test for three different, very important things: We can lyse some of our bacterial culture and try to isolate our his-tagged protein form the lysate. Detection in this step would demonstrate successful expression of our fusion protein. Secondly we can place our bacteria in a solution with high osmolarity. As mentioned this eludes the proteins from the cell wall, which should also work for our fusion proteins. Detection of our protein in this step would demonstrate correct expression as well as localization in the cell wall. Last of all we could place the cells in medium containing the elastase or a protease with the same or very similary specificity. If we are able to pull down the his-tagged peptide then it would demonstrate correct expression, localization and that the cleavage site is accessible to the protease and ComC can be successfully cleaved off.
Detection via Parasite proteases
In order to detect schistosoma in water they have to release their elastse. This does not usually occur in the absense of a stimulus however it is easy to trigger this behaviour in vivo. Upon detection of human (or mice) skin lipids and temperatures close to 37°C, the invasive behaviour is triggered. Several proteases are released from pre- and postacetabular glands or the cercaria at the leading edge of the invading parasite. While multiple enzymes are released, only one protease activity was found to be necessary for invasion in S. mansoni, haematobium and douthitti ([http://books.google.co.uk/books?id=JL7qs-O3z1MC&pg=PA299&lpg=PA299&dq=advances+in+parasitology+london&source=bl&ots=b8mvkz6zyC&sig=t7Pwy0egEDQ9dW-GIZEK7Pb6xxs&hl=en&ei=n05ATKLnJsqNjAePyOAG&sa=X&oi=book_result&ct=result&resnum=3&ved=0CCEQ6AEwAg#v=onepage&q=advances%20in%20parasitology%20london&f=false Kasny et al 2009]): Schistosoma elastase 2a and b (SmCE-1a/b) ([http://www.jbc.org/content/277/27/24618.abstract Salter et al. 2002]). SmCE are trypsin family serine proteases the specificity of which has been investigated by various studies. It appears that the following sites are preferred:
- P4 - S,T
- P3 - S,W,Y
- P2 - P
- P1 - L
Subtle differences exist between SmCE-2a and b as far as their P4 and P3 sites are concerned. Cleavage kinetics were determined for four different sites, P4-P1: SWPL, TWPL, RWPL, RRPL with R previously determined as unfavourable at P4 and P3. For P4 an 11-fold difference in activity was determined between favourable S/T and R, while a 3 fold difference was determined for P3 W to R [http://www.jbc.org/content/277/27/24618.abstract (Salter et al. 2002)]. The most favourable sequence would therefore be SWPL.
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 12:33, 18 October 2010 | 100×100 (5 KB) | Bm108 (Talk | contribs) |
File links
There are no pages that link to this file.
 "
"