Team:Imperial College London/Modelling/Protein Display/Detailed Description
From 2010.igem.org
m |
(Putting in frames for pictures and buttons for dropped stuff. Graphs still need updating) |
||
| Line 10: | Line 10: | ||
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Surface Protein Model | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Surface Protein Model | ||
|- | |- | ||
| - | |<html>This model consists of 5 parts that had to be developed: | + | |<html> |
| + | <head> | ||
| + | <script type="text/javascript" src="http://ajax.googleapis.com/ajax/libs/jquery/1.4.2/jquery.min.js"></"></script> | ||
| + | <script type="text/javascript"> | ||
| + | $(document).ready(function() { | ||
| + | |||
| + | $('div.accordionButton').click(function() { | ||
| + | |||
| + | $(this).next().slideToggle('normal'); | ||
| + | }); | ||
| + | |||
| + | $("div.accordionContent").hide(); | ||
| + | |||
| + | }); | ||
| + | |||
| + | </script> | ||
| + | <style type="text/css"> | ||
| + | #wrapper { | ||
| + | width:850px; | ||
| + | margin-left: auto; | ||
| + | margin-right: auto; | ||
| + | } | ||
| + | |||
| + | .accordionButton { | ||
| + | width: 400px; | ||
| + | margin-left: 200px; | ||
| + | height: 20px; | ||
| + | float: left; | ||
| + | text-align: center; | ||
| + | display: block; | ||
| + | color: #ffffff; | ||
| + | background: #Ea8828; | ||
| + | cursor: pointer; | ||
| + | padding: 10px; | ||
| + | } | ||
| + | |||
| + | .accordionContent { | ||
| + | width: 825px; | ||
| + | float: left; | ||
| + | text-align: justify; | ||
| + | background: #eeeeee; | ||
| + | display: none; | ||
| + | padding: 10px; | ||
| + | } | ||
| + | |||
| + | .accordionButton:hover { | ||
| + | color: #555555; | ||
| + | border-bottom: 1px solid #555555; | ||
| + | } | ||
| + | |||
| + | </style> | ||
| + | </head> | ||
| + | |||
| + | This model consists of 5 parts that had to be developed: | ||
<ol> | <ol> | ||
<li>Identification of all active and relevant elements of the isolated part of the system.</li> | <li>Identification of all active and relevant elements of the isolated part of the system.</li> | ||
| Line 39: | Line 92: | ||
The optimal peptide concentration required to activate ComD is 10 ng/ml <a href="http://ukpmc.ac.uk/backend/ptpmcrender.cgi?accid=PMC40587&blobtype=pdf">[1]</a>. This is the threshold value for ComD activation. However, the minimum concentration of peptide to give a detectable activation is 0.5ng/ml. | The optimal peptide concentration required to activate ComD is 10 ng/ml <a href="http://ukpmc.ac.uk/backend/ptpmcrender.cgi?accid=PMC40587&blobtype=pdf">[1]</a>. This is the threshold value for ComD activation. However, the minimum concentration of peptide to give a detectable activation is 0.5ng/ml. | ||
<br /> | <br /> | ||
| - | The threshold for the minimal activation of the receptor is c<sub>th</sub>=4.4658x10<sup>-9</sup> mol/L. | + | The threshold for the minimal activation of the receptor is c<sub>th</sub>=4.4658x10<sup>-9</sup> mol/L. Click on the button below to uncover the calculations. |
| + | |||
| + | <div id="wrapper"> | ||
| + | <div class="accordionButton">Converting 10 ng/ml to 4.4658x10<sup>-9</sup> mol/L</div> | ||
| + | <div class="accordionContent"> | ||
<ul> | <ul> | ||
<li>The mass of a peptide is 2.24kDa = 3.7184x10<sup>-21</sup>g.</li> | <li>The mass of a peptide is 2.24kDa = 3.7184x10<sup>-21</sup>g.</li> | ||
| Line 46: | Line 103: | ||
<li>The threshold for minimal activation of receptor is 2.2329x10<sup>-10</sup> mol/L.</li> | <li>The threshold for minimal activation of receptor is 2.2329x10<sup>-10</sup> mol/L.</li> | ||
</ul> | </ul> | ||
| + | </div> | ||
| + | </div> | ||
<br/><br/> | <br/><br/> | ||
| Line 54: | Line 113: | ||
This control volume is considered to be wrong, but the details were kept for reference.<br/> | This control volume is considered to be wrong, but the details were kept for reference.<br/> | ||
| + | <br /> | ||
| + | <div id="wrapper"> | ||
| + | <div class="accordionButton">Initial Choice of Control Volume</div> | ||
| + | <div class="accordionContent"> | ||
| + | <b>Control volume initial choice</b><br/> | ||
| + | This control volume is considered to be wrong by us, but the details were kept for the reference.<br/> | ||
The control volume: | The control volume: | ||
| - | The inner boundary is determined by the | + | The inner boundary is determined by the bacterial cell (proteins after being displayed and cleaved cannot diffuse back into bacterium). The outer boundary is more time scale dependent. We have assumed that after mass cleavage of the display-proteins by TEV, many of these AIPs will bind to the receptors quite quickly (eg. 8 seconds). Our volume is determined by the distance that AIPs could travel outwards by diffusion within that short time. In this way, we are sure that the concentration of AIPs outside our control volume after a given time is approximately 0. |
| - | This approach is not very accurate and can lead to false negative conclusions (as in reality there will be a concentration gradient, with | + | This approach is not very accurate and can lead us to false negative conclusions (as in reality there will be a concentration gradient, with highest conentration on the cell wall). |
<br /> | <br /> | ||
</html> | </html> | ||
| - | + | <div ALIGN=CENTER> | |
| + | {| style="background:#e1e1e1;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;" cellspacing="5"; | ||
| + | |- | ||
| + | |<html> | ||
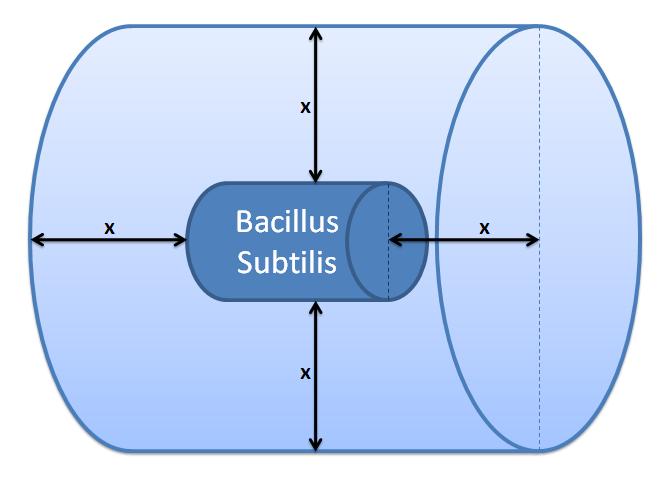
| + | <img src="http://www.openwetware.org/images/4/42/Outer_Volume.JPG" width="300" alt="Control volume (Volume of B.sub to be excluded. x indicates the distance travelled by AIPs fromthe surface in time t)." /> | ||
| + | </html> | ||
| + | |- | ||
| + | |Control volume (Volume of B.sub to be excluded.<br/> x indicates the distance travelled by AIPs from<br/> the surface in time t). | ||
| + | |} | ||
| + | </div> | ||
<html> | <html> | ||
<br /> | <br /> | ||
| Line 87: | Line 161: | ||
<br /> | <br /> | ||
</html> | </html> | ||
| - | + | <div ALIGN=CENTER> | |
| + | {| style="background:#e7e7e7;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;" cellspacing="5"; | ||
| + | |- | ||
| + | |<html> | ||
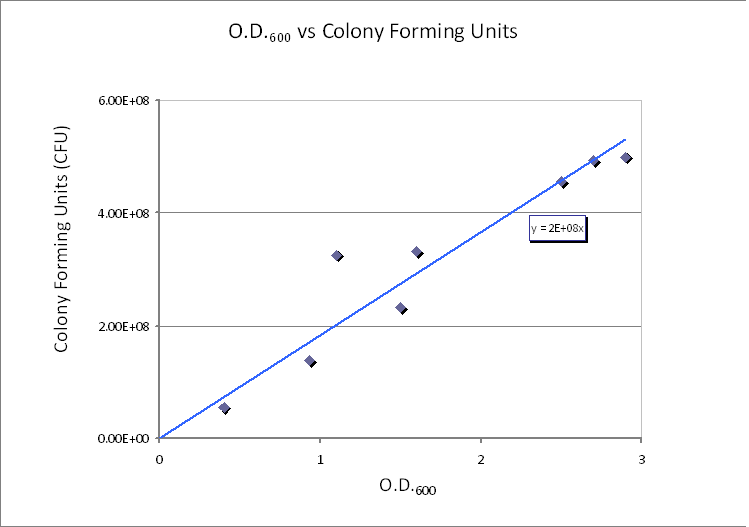
| + | <img src="http://www.openwetware.org/images/b/bf/Calibration-Dilution.PNG" width="600" alt="CFU/ml vs. Optical Density at 600nm (OD600)." /> | ||
| + | </html> | ||
| + | |- | ||
| + | |CFU/ml vs. Optical Density at 600nm (OD600). | ||
| + | |} | ||
| + | </div> | ||
<html> | <html> | ||
<br /> | <br /> | ||
| Line 102: | Line 185: | ||
<br /> | <br /> | ||
</html> | </html> | ||
| - | + | <div ALIGN=CENTER> | |
| + | {| style="background:#e7e7e7;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;" cellspacing="5"; | ||
| + | |- | ||
| + | |<html> | ||
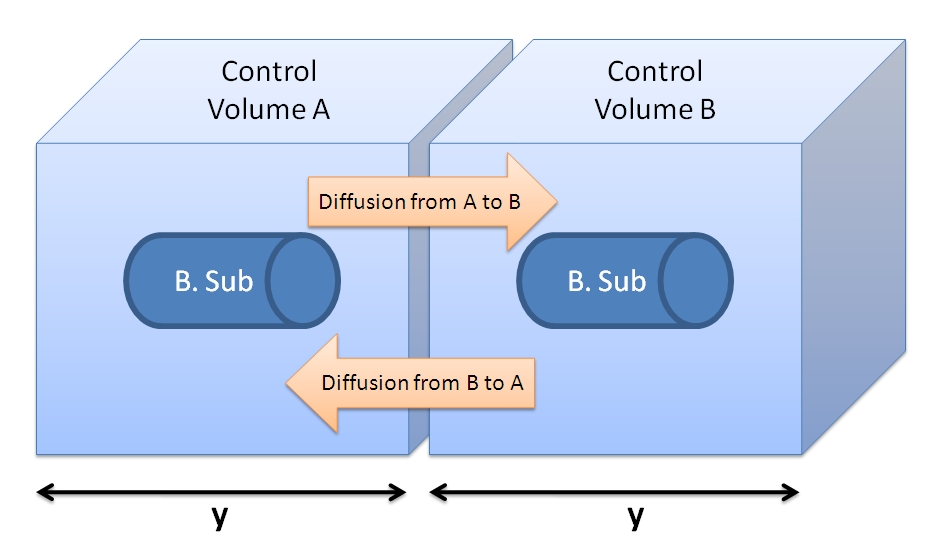
| + | <img src="http://www.openwetware.org/images/f/f9/Control_Volume_and_fluxes.JPG" width="400" alt="Figure showing 2 cells with their control volumes." /> | ||
| + | </html> | ||
| + | |- | ||
| + | |Figure showing two cells with their control volumes. | ||
| + | |} | ||
| + | </div> | ||
<html> | <html> | ||
<br /><br /> | <br /><br /> | ||
| Line 114: | Line 206: | ||
It is not possible to increase the cell density by more than 10<sup>9</sup> CFU/ml</math> because of the size of a cell. | It is not possible to increase the cell density by more than 10<sup>9</sup> CFU/ml</math> because of the size of a cell. | ||
| - | <h2>5. Protein production | + | <h2>5. Protein production</h2> |
<ul> | <ul> | ||
<li>The paper mentions that each cell displays 2.4x10<sup>5</sup> peptides. <a href="http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2000.tb09188.x/pdf">[2]</a></li> | <li>The paper mentions that each cell displays 2.4x10<sup>5</sup> peptides. <a href="http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2000.tb09188.x/pdf">[2]</a></li> | ||
| Line 130: | Line 222: | ||
<br /> | <br /> | ||
</html> | </html> | ||
| - | + | <div ALIGN=CENTER> | |
| + | {| style="background:#e7e7e7;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;" cellspacing="5"; | ||
| + | |- | ||
| + | |<html> | ||
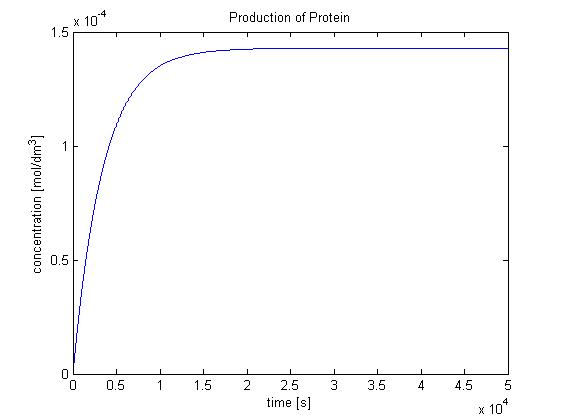
| + | <img src="http://www.openwetware.org/images/f/f5/Protein_production.jpg" width="600" alt="Production of protein by transcription and translation." /> | ||
| + | </html> | ||
| + | |- | ||
| + | |Production of protein by transcription and translation. | ||
| + | |} | ||
| + | </div> | ||
| + | <html> | ||
| + | <br /> | ||
| + | <p>The degradation rate was kept constant, and the production rate was changed according to the final concentration.</p> | ||
| + | |||
| + | |||
| + | <b>Protein production in Control Volume</b><br/> | ||
| + | The previously determined constants of protein production in B.sub to obtain the concentration of proteins are not valid in the Control Volume. It has to be adjusted (multiplied) by the following factor: | ||
| + | <br /> | ||
| + | factor=V<sub>bacillus</sub>/V<sub>CV</sub> = 5.7974x10<sup>-6</sup> (for the particular numbers presented above) | ||
| + | </html> | ||
| + | <div ALIGN=CENTER> | ||
| + | {| style="background:#e7e7e7;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;" cellspacing="5"; | ||
| + | |- | ||
| + | |<html> | ||
| + | <img src="http://www.openwetware.org/images/7/78/Protein_in_control_volume.jpg" width="600" alt="Production of protein by transcription and translation in control volume." /> | ||
| + | </html> | ||
| + | |- | ||
| + | |Production of protein by transcription and translation in control volume. | ||
| + | |} | ||
| + | </div> | ||
<html> | <html> | ||
<h2>References</h2> | <h2>References</h2> | ||
Revision as of 15:17, 20 October 2010
| Temporary sub-menu: Objectives; Detailed Description; Parameters & Constants; Results & Conclusion;Download MatLab Files; |
| Surface Protein Model | ||||||||||
This model consists of 5 parts that had to be developed:
1. Elements of the system
2. Interactions between elementsApart from the proteins being expressed from genes, there was only one more chemical reaction identified in this part of the system. This is the cleavage of proteins, which is an enzymatic reaction:
This enzymatic reaction can be rewritten as partial differential equations (PDEs), which is of similar form as the 1-step amplification model. However, most of the constants and initial concentrations are different. For detailed description and derivation of PDEs, please refer to "Detailed Description" part of Modelling Output. 3. Threshold concentration of AIPThe optimal peptide concentration required to activate ComD is 10 ng/ml [1]. This is the threshold value for ComD activation. However, the minimum concentration of peptide to give a detectable activation is 0.5ng/ml.The threshold for the minimal activation of the receptor is cth=4.4658x10-9 mol/L. Click on the button below to uncover the calculations. Converting 10 ng/ml to 4.4658x10-9 mol/L
4. Control volume selectionNote that this enzymatic reaction is modelled outside the cell. Hence, it is important to take into account the cell boundaries. It is worth considering whether diffusion or fluid movements will play a significant role. Initially, we defined a control volume assuming that bacteria would grow in close colonies on the plate. This control volume is considered to be wrong, but the details were kept for reference.Initial Choice of Control Volume
Control volume initial choice
This control volume is considered to be wrong by us, but the details were kept for the reference. The control volume: The inner boundary is determined by the bacterial cell (proteins after being displayed and cleaved cannot diffuse back into bacterium). The outer boundary is more time scale dependent. We have assumed that after mass cleavage of the display-proteins by TEV, many of these AIPs will bind to the receptors quite quickly (eg. 8 seconds). Our volume is determined by the distance that AIPs could travel outwards by diffusion within that short time. In this way, we are sure that the concentration of AIPs outside our control volume after a given time is approximately 0. This approach is not very accurate and can lead us to false negative conclusions (as in reality there will be a concentration gradient, with highest conentration on the cell wall).
We realized that our initial choice of control volume was not accurate because we had assumed that the bacteria was the medium. However, in reality bacteria live in colonies very close to each other. Since our bacteria was meant to be used in suspension we had to reconsider this issue. Using CFU to estimate the spacing between cells CFU stands for Colony-forming unit. It is a measure of bacterial numbers. For liquids, CFU is measured per ml. We already have data of CFU/ml from the Imperial iGEM 2008 team, so we could use this data to estimate the number of cells in a given volume using a spectrometer at 600nm wavelength. The graph below is taken from the Imperial iGEM 2008 Wiki page [4].
Side length of CV = y = 1.26x10-4 dm = 1.26x10-5 m. Choice of Control Volume allows simplifications
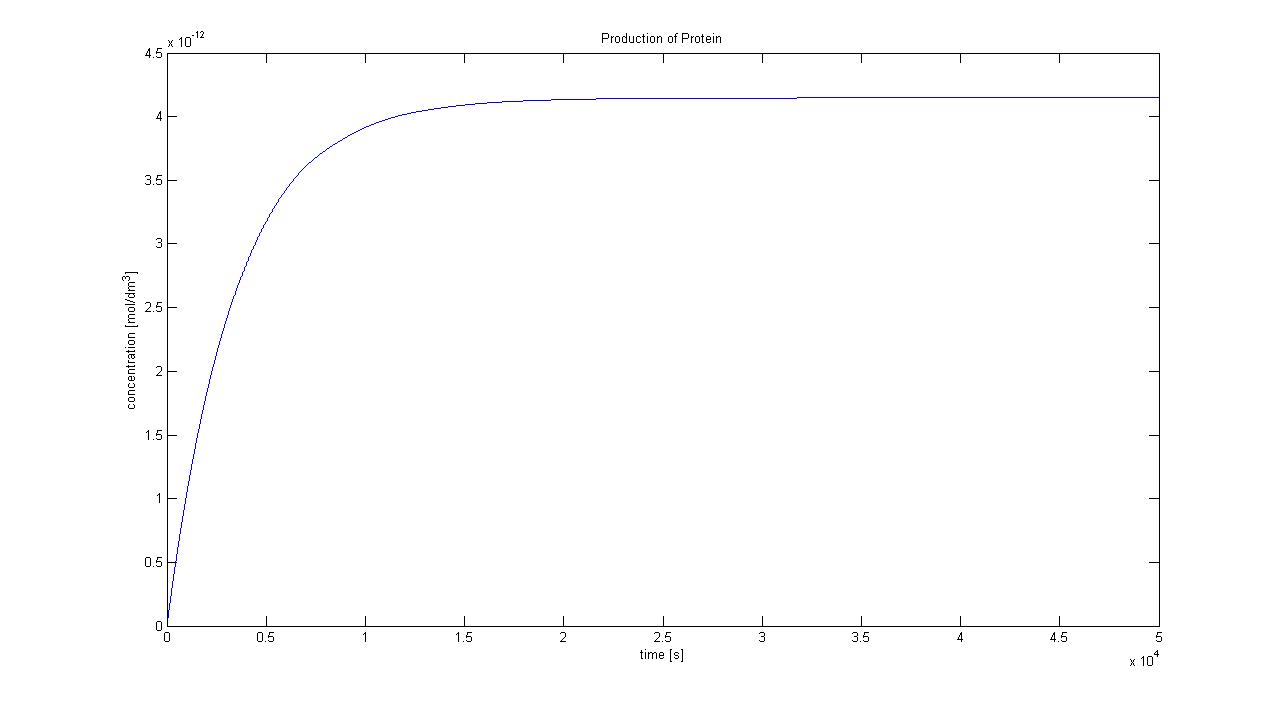
5. Protein production
Hence, we can deduce that the final concentration that the protein expression will tend to is: c = 1.4289x10-4 mol/dm3 = cfinal. Therefore, we can model the protein production by transcription and translation and adjust the production constants so that the concentration will tend towards cfinal. Using a similar model to the simple production of Dioxygenase for the Output Amplification Model (Model preA), we obtain the following graph:
The degradation rate was kept constant, and the production rate was changed according to the final concentration. Protein production in Control VolumeThe previously determined constants of protein production in B.sub to obtain the concentration of proteins are not valid in the Control Volume. It has to be adjusted (multiplied) by the following factor: factor=Vbacillus/VCV = 5.7974x10-6 (for the particular numbers presented above)
References
|
 "
"