Switch
From 2010.igem.org
(→The switches) |
|||
| (8 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
<!-- ############## WIKI-PAGE STARTS HERE ############## --> | <!-- ############## WIKI-PAGE STARTS HERE ############## --> | ||
| - | == | + | =concept= |
| - | Our switches are based on the principle of transcriptional antitermination. Transcription can be cancelled by the formation of a RNA stem loop in the nascent RNA-chain, which then causes the RNA polymerase to stop transcription and fall off the DNA. We use sequences, that are capable of stem loop formation, | + | |
| + | ==bioLOGICS== | ||
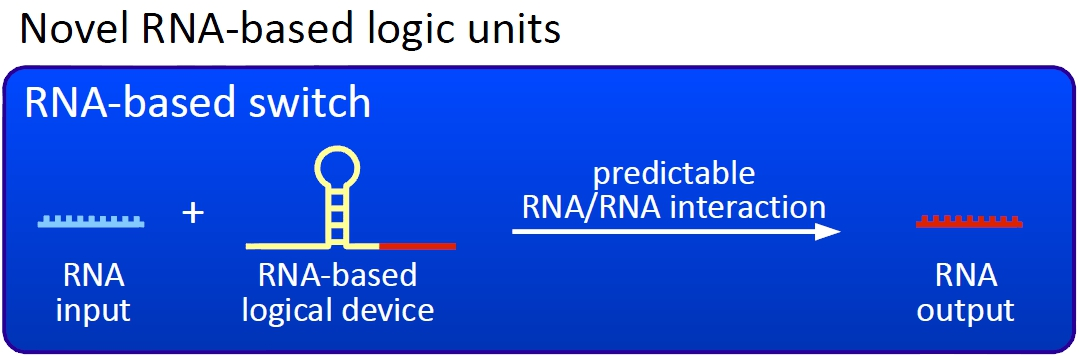
| + | Our switches are based on the principle of transcriptional antitermination. Transcription can be cancelled by the formation of a RNA stem loop in the nascent RNA-chain, which then causes the RNA polymerase to stop transcription and fall off the DNA. We use sequences as transcriptional switches, that are calculated to be capable of stem loop formation.<br> | ||
| + | [[Image:TUM2010 novelunit.png|thumb|right|300 px|Novel switches based on RNA-RNA interaction]] | ||
| + | |||
| + | We plan to control the termination at these switches with a small RNA molecule (our RNA "signal") that is complementary to a part of the stem loop forming sequence. This small functional RNA inhibits the stem loop formation by complementary base-pairing and hence avoids transcription termination. The signal is composed of two parts: While the first part provides specifity, the second part causing stem loop disintegration can in principle be the same for all bioLOGICS. Therefore variation of the first part allows the construction of an endless number of switches. The second part causing stem loop disintegration is based on a working system (see attenuation) established by nature. Different stem loops were tested in this effort: <font color="red">Regulatory parts from the E. coli ''trp''-operon, ''his''-operon and one based on previous iGEM-work.</font> | ||
<br> | <br> | ||
| - | + | The initial signal can be provided by various metabolic compounds and stimuli and the output signal can be anything DNA-coded, too. In the last years, many working sensory systems were submitted to the Partsregistry. Those parts can now be utilized as inputs for our network. BioLOGICS provides a new way to use the whole potential of iGEM distributions connecting different parts for totally new applications. | |
<br> | <br> | ||
| - | + | The major advantage over conventional protein based devices and even [[glossary|riboswitches]] is the small size of our basic units, its easy construction, huge variability and easy upscaling to complex networks in cells. Furthermore introduction of an RNA network into ''E. coli'' is especially easy - all informations can be coded on one plasmid. In comparison to protein networks, the elegance of RNA based networks lies in its functionality, simplicity and predictability. Since RNA-RNA interactions are highly predictable and since our system is based on the variation of one principle in many switches, simulations of our networks are much more accurate than those of comparable systems currently available. Incorporation of established Biobricks allows a high diversity of input and output signals providing a whole new level of cell control. | |
| - | + | ||
<div align="justify"></div> | <div align="justify"></div> | ||
| - | + | <!-- | |
Note, that our systems work with termination on transcription level, so in our case the RNA-Polymerase falls off instead of stalling a ribosome. Antitermination means the avoidance of trancription termination by interaction with another RNA sequence (the '''signal'''), which binds to the transcription mRNA and anticipates the formation of a stem loop. So basicly one gets a yes/no answer with a first '''switch''': If the small RNA piece is available, transcription will continue. If not, it will be terminated. This is the way our first devices should work. <br> | Note, that our systems work with termination on transcription level, so in our case the RNA-Polymerase falls off instead of stalling a ribosome. Antitermination means the avoidance of trancription termination by interaction with another RNA sequence (the '''signal'''), which binds to the transcription mRNA and anticipates the formation of a stem loop. So basicly one gets a yes/no answer with a first '''switch''': If the small RNA piece is available, transcription will continue. If not, it will be terminated. This is the way our first devices should work. <br> | ||
Latest revision as of 13:23, 10 October 2010
|
||||||||
|
|
conceptbioLOGICSOur switches are based on the principle of transcriptional antitermination. Transcription can be cancelled by the formation of a RNA stem loop in the nascent RNA-chain, which then causes the RNA polymerase to stop transcription and fall off the DNA. We use sequences as transcriptional switches, that are calculated to be capable of stem loop formation. We plan to control the termination at these switches with a small RNA molecule (our RNA "signal") that is complementary to a part of the stem loop forming sequence. This small functional RNA inhibits the stem loop formation by complementary base-pairing and hence avoids transcription termination. The signal is composed of two parts: While the first part provides specifity, the second part causing stem loop disintegration can in principle be the same for all bioLOGICS. Therefore variation of the first part allows the construction of an endless number of switches. The second part causing stem loop disintegration is based on a working system (see attenuation) established by nature. Different stem loops were tested in this effort: Regulatory parts from the E. coli trp-operon, his-operon and one based on previous iGEM-work.
|
|||||||
 "
"