Team:WashU/Notebook/Yeast
From 2010.igem.org
(Difference between revisions)
BrianLandry (Talk | contribs) (→7/15) |
(→2010/08/18) |
||
| (14 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{WashUHeader}} | {{WashUHeader}} | ||
| - | == | + | ==2010/06/24== |
| + | *plated Yeast Gal-YFP MATa, trp1, leu2::GAL1-YFP URA3/Kanr and Yeast YF4 MATa Prototroph on a Kan plate and a YPD plate | ||
| + | *Started a liquid culture of Yeast YF4 MATa Prototroph in YPD | ||
| + | <br> | ||
| - | == | + | ==2010/06/29== |
| - | + | *A practice transformation was preformed at the Cohen Lab with Ilaria. | |
| - | + | *The OD600 was 0.651 which was low, but used in order to speed up the experiment. | |
| + | *The PMKV005 loxKanlox Plasmid (McCusker 2005 Yeast, 21: 163-171) was used into transform wildtype yeast. | ||
| + | *The plates used were Kan positive plates created on 6/24. | ||
| + | *However it was later found that these plates might not correctly select for drug resistance. | ||
| + | *Two plates were made, one with the transformant and one with a control. | ||
| + | *The attached procedure was used and annotated during the experiment | ||
| + | <br> | ||
| - | + | ==2010/07/01== | |
| + | *Examined the Sample and Control plates from the practice transformation on June 29th. | ||
| + | *Two colonies had grown on the Sample plate and none showed on the control plate. | ||
| + | *The low colony number could be due to a low OD600 (0.651) used in the transformation. | ||
| + | *Plated the two colonies on a G418 plate in order to conduct the first round of purification | ||
| + | <br> | ||
| - | == | + | ==2010/07/02== |
| + | *PRACTICE YEAST TRANSFORMATION: | ||
| + | **Colonies grew on the first purification plate for both colony 1 and 2. | ||
| + | **Two colonies were plated from both transformed colony 1 and transformed colony 2 onto a second and final purification plate | ||
| - | + | *PLATES: | |
| - | + | YPD: all yeast grew | |
| + | His-: only 1&2 (WT) yeast grew, (4,6,11,12 did not grow) | ||
| + | Kan:3,4,7 grew- 1&2 were selected against but some colonies were visible | ||
| + | Ura-: only 1&2 (WT) yeast grew, (11 did not grow) | ||
| - | + | *Amp: bacteria 8,9,10 grew (other amp plate WT control did not grow) | |
| - | + | ||
| - | + | ||
| - | + | *Conclusion: all the resistance plates work in that they select against those w/o the resistance; however, there was some low level of growth on some of these plates where there was not supposed to be | |
| - | + | <br> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ==2010/07/12== | |
| + | *Streaked out wild type yeast strain FY4 onto a YPD plate and inoculated about 5 mL of YPD- incubate and shake | ||
| + | *Brian: The FY5 freezer stock was accidentally allowed to thaw and therefor a second freezer stock was started by growing an overnight culture inoculated from the original freezer stock. | ||
| + | *The freezer stock was made according to protocol from the cohen lab. | ||
| + | <br> | ||
| + | ==2010/07/13== | ||
| + | *Plated FY4 yeast onto a YPD plate to grow up for a practice transformation in case the plate from the freezer stock does not grown since there were no visible growths on it. | ||
| + | *The FY4 was taken from a Ura- plate plated on 6/29 because it had the best colonies out of all the plates in the refrigerator. | ||
| + | *A freezer stock of FY4 was made according to protocol from an overnight culture of the FY4 strain since the original stock was destroyed when plating on 7/12. | ||
| + | <br> | ||
| - | + | ==2010/07/14== | |
| + | *Yeast colonies were observed on the FY4 plated on 7/12 from the FY4 freezer stock. This plate was brought to the Cohen lab along with liquid YPD-4 and they were used to inoculate a 5ml overnight culture. | ||
| + | <br> | ||
| - | == | + | ==2010/07/15== |
| - | + | *Yeast Transformation | |
| - | + | *Alice and Brian went to the Cohen Lab to preform a yeast transformation. | |
| - | + | *Ilaria showed us how to use the nano-drop using the following procedure: | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | Yeast Transformation | + | |
| - | Alice and Brian went to the Cohen Lab to preform a yeast transformation. Ilaria showed us how to use the nano-drop using the following procedure: | + | |
1. Wipe the nanodrop with a kim-wipe | 1. Wipe the nanodrop with a kim-wipe | ||
2. Click the shortcut on the computer that is identicle to the logo on the nano drop | 2. Click the shortcut on the computer that is identicle to the logo on the nano drop | ||
| Line 57: | Line 66: | ||
8. Put on 1 μL of plasmid | 8. Put on 1 μL of plasmid | ||
9. Make sure sample type is set to DNA 50 | 9. Make sure sample type is set to DNA 50 | ||
| - | Looked at the graph, a spike at 130 is due to salts, a spike at 260 is due to DNA, a spike at 280 is due to protein. Record the 260/230 ratio and 260/280 ratios, you want then both to be above 1, somewhere between 1-2 is good | + | *Looked at the graph, a spike at 130 is due to salts, a spike at 260 is due to DNA, a spike at 280 is due to protein. *Record the 260/230 ratio and 260/280 ratios, you want then both to be above 1, somewhere between 1-2 is good |
| - | A DNA concentration of ~1ng/ μL was measured which is extremely low, and therefor the plasmid should not be used to preform a yeast transformation. This could be a poissible explination for the extremely low transformation efficiency of Transformation-1. Because of this it is necessary to amplify the plasmid in E. coli. | + | *A DNA concentration of ~1ng/ μL was measured which is extremely low, and therefor the plasmid should not be used to preform a yeast transformation. |
| - | Amplification of PMKV005 | + | *This could be a poissible explination for the extremely low transformation efficiency of Transformation-1. |
| - | The PMKV005 was to be amplified in E. coli to increase its concentration. Vorachek-Warren | + | *Because of this it is necessary to amplify the plasmid in ''E. coli''. |
| - | et. al. 2004 showed that PMKV005 was derived from the pAG26 plasmid which had ampicillin resistance. | + | *Amplification of PMKV005 |
| - | The competent cells were obtained from NEB thawed on ice and transformed with the following protocol. | + | **The PMKV005 was to be amplified in ''E. coli'' to increase its concentration. |
| + | **Vorachek-Warren et. al. 2004 showed that PMKV005 was derived from the pAG26 plasmid which had ampicillin resistance. | ||
| + | **The competent cells were obtained from NEB thawed on ice and transformed with the following protocol. | ||
1. Pipette 1 μL of plasmid into the cells. Flick the tubes very gently; | 1. Pipette 1 μL of plasmid into the cells. Flick the tubes very gently; | ||
2. Let them site on ice for 10 minutes; | 2. Let them site on ice for 10 minutes; | ||
| Line 71: | Line 82: | ||
7. Plate 50 μL on a antibiotic resistant plate (which you have warmed up with the lid slightly open for about 20 minutes); | 7. Plate 50 μL on a antibiotic resistant plate (which you have warmed up with the lid slightly open for about 20 minutes); | ||
8. incubate overnight at 37 degC. | 8. incubate overnight at 37 degC. | ||
| + | <br> | ||
| - | == | + | ==2010/07/16== |
| - | Plated a colony from the PMKV005 e. coli transformation on 7/15 into LB for an overnight culture. | + | *Plated a colony from the PMKV005 e. coli transformation on 7/15 into LB for an overnight culture. |
| - | + | <br> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | == | + | ==2010/07/17== |
| - | + | *Conducted a Miniprep from the o/n culture of PMKV005 transformed ''E. coli'' made on 7/16. | |
| + | *Conducted using the Plasmid DNA Purification Using the QIAprep Spin Miniprep Kit and a Microcentrifuge protcol on page 22 of the attached manual. | ||
| + | *All places when centrifugation is between 30-60 seconds 60 seconds were used. | ||
| + | *The 3 LB plates from the Mol. Bio. group were also examined and no colonies were observed. | ||
| - | == | + | ==2010/07/18== |
| + | *Made an overnight culture of the FY4 strain using YPD-2 | ||
| + | <br> | ||
| - | == | + | ==2010/07/19== |
| - | + | *Prepared an o/n culture of FY4 strain from plate made on 7/12 for transformation on 7/20 | |
| - | + | *Nanodropped the PMKV005 plasmid purified on 7/17 and found a concentration of 22.8 μL | |
| - | + | *Conducted a Yeast Transformation following the protocol received from Ilaria | |
| - | FY4 cells were used from culture started on 7/18. | + | *FY4 cells were used from culture started on 7/18. |
| - | + | *Cell inoculated at 8:55am and put in shaker at 37 ° C until 12:30pm when it was transported over to the Cohen lab. OD600 taken at 12:50 measure 0.318. At 2:45 a final OD600 of 0.631 was reached. | |
| - | + | *The T-mix was prepared x6 and the salmon sperm was boiled at x9 because some boils off | |
| - | 9,10. The cells were transformed with the following mixtures. | + | *9,10. The cells were transformed with the following mixtures. |
| - | Vial # Concentration of DNA (ng/μL) Volume of DNA (μL) Mass of DNA (ng) Volume of T-mix | + | Vial # Concentration of DNA (ng/μL) Volume of DNA (μL) Mass of DNA (ng) Volume of T-mix |
1 22.8 0.877 20 359 | 1 22.8 0.877 20 359 | ||
2 22.8 0.439 10 359.5 | 2 22.8 0.439 10 359.5 | ||
| Line 100: | Line 113: | ||
5 0 0 0 360 | 5 0 0 0 360 | ||
| - | Our first round of Kan Plates were used to plate the colonies. | + | *Our first round of Kan Plates were used to plate the colonies. |
| + | <br> | ||
| + | ==2010/07/20== | ||
| + | *Attempted to conduct a transformation back at our lab. | ||
| + | *However we were unable to get an OD600 reading. | ||
| + | *The nano-drop gave inconsistent and negligible results. | ||
| + | *Using a spectrophotometer an OD of just 0.004 was recorded, on two separate trials. | ||
| + | *The following procedure was used for the spectrophotometer. | ||
| + | *An o/n culture of the FY4 strain plated on 7/12 was made. | ||
| - | + | 1. Turn on the instrument by rotating the 0%T dial clockwise. | |
| - | + | ||
| - | + | ||
| - | 1.Turn on the instrument by rotating the 0%T dial clockwise. | + | |
You should hear it click when it turns on. | You should hear it click when it turns on. | ||
| - | 2.Allow the spec to warm up for at least 15 minutes. | + | 2. Allow the spec to warm up for at least 15 minutes. |
| - | Given the long warm up time we will likely be leaving this device on during periods of heavy use | + | Given the long warm up time we will likely be leaving this device on during periods of heavy use. |
| - | 3.After the warm-up period, set the wavelength to 600 nm (top knob). | + | 3. After the warm-up period, set the wavelength to 600 nm (top knob). |
| - | 4.Set the filter lever to the appropriate position for the selected wavelength (bottom). | + | 4. Set the filter lever to the appropriate position for the selected wavelength (bottom). |
| - | 5.Press the MODE button until the display mode is set to FACTOR. | + | 5. Press the MODE button until the display mode is set to FACTOR. |
| - | 6.Using the INCREASE/DECREASE buttons ensure that the factor is set to 1.0. | + | 6. Using the INCREASE/DECREASE buttons ensure that the factor is set to 1.0. |
| - | 7.Press the MODE button until the display mode is set to TRANSMITTANCE. | + | 7. Press the MODE button until the display mode is set to TRANSMITTANCE. |
| - | 8.With no cuvette in the holder and the lid closed, turn the 0%T knob until the measured transmittance is 0.0. | + | 8. With no cuvette in the holder and the lid closed, turn the 0%T knob until the measured transmittance is 0.0. |
| - | 9.Fill a cuvette with 3.0 mL of the medium used to grow the cells (e.g. YPD) and wipe the sides with a KimWipe to eliminate fingerprints and dust particles. | + | This is zeroing the device. |
| - | 10.Place the cuvette in the compartment. | + | 9. Fill a cuvette with 3.0 mL of the medium used to grow the cells (e.g. YPD) and wipe the sides with a KimWipe to eliminate fingerprints and dust particles. |
| - | 11.Align the guide mark of the cuvette with the guide mark on the compartment. | + | 10. Place the cuvette in the compartment. |
| - | 12.Close the lid of the compartment. | + | 11. Align the guide mark of the cuvette with the guide mark on the compartment. |
| - | 13.Adjust the TRANSMITTANCE to 100.0 using the 100%T knob (right). | + | 12. Close the lid of the compartment. |
| - | + | 13. Adjust the TRANSMITTANCE to 100.0 using the 100%T knob (right). | |
| - | 14.Remove the sample. | + | This is essentially the equivalent of when you TARE a balance. |
| - | + | 14. Remove the sample. | |
| - | 15.In a new cuvette add 3.0 mL of your cell culture sample. | + | You may choose to leave this cuvette of YPD for other users as they will likely be performing the same procedure. |
| - | 16.Wipe the cell with a KimWipe and insert into the compartment as in Step 10. | + | Note: the spectrophotometer must be blanked for each user even if a common cuvette is being used. |
| - | 17.Record the value shown in the data display (make sure MODE is ABSORBANCE). | + | 15. In a new cuvette add 3.0 mL of your cell culture sample. |
| - | + | 16. Wipe the cell with a KimWipe and insert into the compartment as in Step 10. | |
| - | 18.When all measurements are obtained, turn the O%T (left knob) counterclockwise to switch the spec off. | + | 17. Record the value shown in the data display (make sure MODE is ABSORBANCE). |
| - | 19.Clean your cuvettes thoroughly and dry them completely. | + | Absorbance is a synonym for OD (optical density). |
| + | 18. When all measurements are obtained, turn the O%T (left knob) counterclockwise to switch the spec off. | ||
| + | 19. Clean your cuvettes thoroughly and dry them completely. | ||
| - | Taken from http://fg.cns.utexas.edu/fg/course_notebook_chapter_nine.html | + | *Taken from http://fg.cns.utexas.edu/fg/course_notebook_chapter_nine.html |
| - | + | <br> | |
| - | == | + | ==2010/07/21== |
| - | + | *Measured the OD600 of the overnight FY4 culture using a spectrophotometer and a nanodrop | |
| - | + | **Spectrophotometer gave a reading of 1.220 | |
| - | + | **Nano drop gave readings of .282, 0.347, 0.360 | |
| - | The nanodrop has a 1mm pathlength while the spectrophotometer has a 1 cm pathlength so there is an expected factor of 10 difference | + | *The nanodrop has a 1mm pathlength while the spectrophotometer has a 1 cm pathlength so there is an expected factor of 10 difference |
| - | We later became aware that the spectrophotometer cannot measure saturated cultures accurately so we conducted a more thorough experiment. The culture was diluted 5 fold and the absorbance of both the saturated and diluted cultures were measured. | + | *We later became aware that the spectrophotometer cannot measure saturated cultures accurately so we conducted a more thorough experiment. |
| - | + | *The culture was diluted 5 fold and the absorbance of both the saturated and diluted cultures were measured. | |
| + | *Results | ||
Absorbance OD600 | Absorbance OD600 | ||
Spectrophotomer Saturated Culture 1.055 1.055 | Spectrophotomer Saturated Culture 1.055 1.055 | ||
| Line 151: | Line 172: | ||
Nanodrop 1/5 dilution trial 3 0.712 7.12 | Nanodrop 1/5 dilution trial 3 0.712 7.12 | ||
Nanodrop 1/5 dilution trial 4 -0.677 -6.77 | Nanodrop 1/5 dilution trial 4 -0.677 -6.77 | ||
| - | From these results we have chosen not to use the nanodrop to measure OD600 of cell cultures because it gives very sporadic readings instead the spectrophotometer will be used. | + | *From these results we have chosen not to use the nanodrop to measure OD600 of cell cultures because it gives very sporadic readings instead the spectrophotometer will be used. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | == | + | *An overnight culture of the FY4 strain was started using YPD-1 |
| - | YPD: The YPD made in the cohen lab on 7/19 was divided up into separate bottles | + | *The FY5 strain was plated from freezer stock and from the plate with strains 1-12 on it |
| - | Transformation: A culture was started using 50 ml of the remainder of the YPD from the first batch and 750 μL of overnight culture. An OD600 reading was taken 6 hours later and it recorded only .8 | + | 3. Gal-YFP |
| - | Growth Curve Analysis: A 50ml culture was started with an OD600 of 0.10 in a 250 ml flask, capped with a medal top and place on the shaker at 100rpm in 30 degC. OD600 readings were taken every hour. | + | 4. Gal-CFP |
| - | + | 5. Control YFP | |
| - | + | 6. Control CFP | |
| + | 7. Gal CFP-YFP | ||
| + | <br> | ||
| + | ==2010/07/22== | ||
| + | *YPD: The YPD made in the cohen lab on 7/19 was divided up into separate bottles | ||
| + | *Transformation: A culture was started using 50 ml of the remainder of the YPD from the first batch and 750 μL of overnight culture. | ||
| + | **An OD600 reading was taken 6 hours later and it recorded only .8 | ||
| + | **Therefore the transformation was aborted due to slow growth. | ||
| + | *Growth Curve Analysis: A 50ml culture was started with an OD600 of 0.10 in a 250 ml flask, capped with a medal top and place on the shaker at 100rpm in 30 degC. | ||
| + | **OD600 readings were taken every hour. | ||
| + | *Overnight culture OD600: 5X dilution OD600=0.392 therefor the OD600 of the overnight culture is 1.69 | ||
| + | *1.96* culture volume/50ml=0.1 therefor the culture volume used was 2.55ml | ||
Time Time Passed OD600 Notes | Time Time Passed OD600 Notes | ||
| Line 181: | Line 205: | ||
19:10:00 9:30:00 2.136 Used 3x dilution | 19:10:00 9:30:00 2.136 Used 3x dilution | ||
23:02:00 13:22:00 4.536 used 6x diltution | 23:02:00 13:22:00 4.536 used 6x diltution | ||
| - | + | <br> | |
| - | == | + | ==2010/07/23== |
| - | Contamination Detection | + | *Contamination Detection |
| - | The overnight culture created on 7/23 was examined under a light microscope for contamination. 100 μL of o/n culture was placed in a microcentrifuge tube and then vortexed to seperate the cells. 10 μL of the culture from the microcentrifuge tube was then placed on a glass slide and covered with a coverslip. | + | **The overnight culture created on 7/23 was examined under a light microscope for contamination. |
| - | + | **100 μL of o/n culture was placed in a microcentrifuge tube and then vortexed to seperate the cells. | |
| - | Conclusions | + | **10 μL of the culture from the microcentrifuge tube was then placed on a glass slide and covered with a coverslip. |
| - | It was concluded that no observable contamination of the cell culture was seen. | + | **The cells were observed at 40x with a Nikon Eclipse 200 microscope. |
| + | **Small circular cells were observed moving across the screen at a slow pace. The cells formed groupings of 2-3. | ||
| + | **Nothing other of note was observed. | ||
| + | *Conclusions: | ||
| + | *It was concluded that no observable contamination of the cell culture was seen. | ||
| - | Yeast Transformation with pMKV005: attempt #2 | + | *Yeast Transformation with pMKV005: attempt #2 |
| - | Overnight culture created on 7/22 was brought to an OD of 0.174 to grow for around 5 hours to a final OD of 0.7. At this time, the yeast was transformed with 20 ng, 2ng, and 0.2 ng of pMKV005 and plated on YPD/Kan plates. A control was also made. No colonies were seen on any of the plates after 2 days of incubation. | + | **Overnight culture created on 7/22 was brought to an OD of 0.174 to grow for around 5 hours to a final OD of 0.7. |
| + | **At this time, the yeast was transformed with 20 ng, 2ng, and 0.2 ng of pMKV005 and plated on YPD/Kan plates. | ||
| + | **A control was also made. No colonies were seen on any of the plates after 2 days of incubation. | ||
| + | <br> | ||
| - | == | + | ==2010/07/26== |
| - | + | *Ran a digest and gel with the mol. bio. group. (All in μL) | |
| - | + | ||
| - | + | ||
| - | Ran a digest and gel with the mol. bio. group. | + | |
| - | All in μL | + | |
1 2 3 4 5 6 7 | 1 2 3 4 5 6 7 | ||
Vector PMKV005 PMKV005 PMKV005 Ksa1 Ksa2 Ksb1 Ksb2 | Vector PMKV005 PMKV005 PMKV005 Ksa1 Ksa2 Ksb1 Ksb2 | ||
| Line 207: | Line 234: | ||
BSA 0.5 0.5 0.5 0.5 0.5 0.5 0.5 | BSA 0.5 0.5 0.5 0.5 0.5 0.5 0.5 | ||
| - | Expected results of PMKV005 digest: | + | *Expected results of PMKV005 digest: |
| - | + | <center> | |
[[Image:WashU_7-27Map.png|400px]] | [[Image:WashU_7-27Map.png|400px]] | ||
| - | + | </center> | |
| - | == | + | <br> |
| - | + | ==2010/07/27== | |
| - | + | *Replated the FY4 plate from 7/26 since the colonies were too dense. | |
| - | + | *Two minipreps were preformed on 2 different cultures that had been transformed with the PMKV005 Plasmid. The Qiagen protocol was followed except when the cells were initially spun down the centrifuge was set to 3000 rpm for 10 minutes. To increase the concentration of one of the samples a 30 μL elution instead of a 50 μL elution was used. | |
| - | The samples were nanodroped following standard protocol and the follow data was obtained: | + | *The samples were nanodroped following standard protocol and the follow data was obtained: |
1: 50 μL elution | 1: 50 μL elution | ||
262.4 ng/μL | 262.4 ng/μL | ||
| Line 225: | Line 252: | ||
260/230=2.30 | 260/230=2.30 | ||
| - | + | *Preformed a Yeast transformation follwing illaria's procedure at our lab. | |
| - | The following conditions were used | + | **The following conditions were used |
1. FY4 Cohen T-Mix 0ng YPD Plate | 1. FY4 Cohen T-Mix 0ng YPD Plate | ||
2. FY4 Cohen T-Mix 0ng G418 Plate | 2. FY4 Cohen T-Mix 0ng G418 Plate | ||
| Line 240: | Line 267: | ||
12. FY5 Our T-Mix 500ng G418 Plate | 12. FY5 Our T-Mix 500ng G418 Plate | ||
13. Cells with G418 resistance G418 Plate | 13. Cells with G418 resistance G418 Plate | ||
| - | The experiment was designed to test the following conditions: | + | *The experiment was designed to test the following conditions: |
| - | Test whether the cells are dieing during the transformation | + | **Test whether the cells are dieing during the transformation |
| - | 1. FY4 Cohen T-Mix 0ng YPD Plate | + | 1. FY4 Cohen T-Mix 0ng YPD Plate |
| - | 7. FY5 Cohen T-Mix 0ng YPD Plate | + | 7. FY5 Cohen T-Mix 0ng YPD Plate |
| - | Test the control to insure transformation is necessary for colony growth | + | *Test the control to insure transformation is necessary for colony growth |
| - | 2. FY4 Cohen T-Mix 0ng G418 Plate | + | 2. FY4 Cohen T-Mix 0ng G418 Plate |
| - | 8. FY5 Cohen T-Mix 0ng G418 Plate | + | 8. FY5 Cohen T-Mix 0ng G418 Plate |
| - | Test whether a G418 Strain can survive on our G418 Plates | + | *Test whether a G418 Strain can survive on our G418 Plates |
| - | 13. Cells with G418 resistance G418 Plate | + | 13. Cells with G418 resistance G418 Plate |
| - | Test the affect of using 500 ng of DNA in place of 100ng of DNA | + | *Test the affect of using 500 ng of DNA in place of 100ng of DNA |
| - | 3. FY4 Cohen T-mix 100ng G418 Plate | + | 3. FY4 Cohen T-mix 100ng G418 Plate |
| - | 4. FY4 Cohen T-Mix 500ng G418 Plate | + | 4. FY4 Cohen T-Mix 500ng G418 Plate |
| - | 9. FY5 Cohen T-Mix 100ng G418 Plate | + | 9. FY5 Cohen T-Mix 100ng G418 Plate |
| - | 10. FY5 Cohen T-Mix 500ng G418 Plate | + | 10. FY5 Cohen T-Mix 500ng G418 Plate |
| - | + | *Experimental Notes | |
| - | Experimental Notes | + | **OD600 Readings |
| - | OD600 Readings | + | |
Time OD600 | Time OD600 | ||
Overnight FY4 0.454X6=2.724* | Overnight FY4 0.454X6=2.724* | ||
| Line 276: | Line 302: | ||
*The first 4 readins were actually taken at wavelength 579nm but this should have very small affect on the values | *The first 4 readins were actually taken at wavelength 579nm but this should have very small affect on the values | ||
| + | *During the heat bath step it was observed that the cells settled at the bottem of the microcentrifuge tubes, we are not sure if this is normal or what affect it would have on the experiment. | ||
| + | <br> | ||
| + | ==2010/07/28== | ||
| + | *YPD and G418 plates were started at the cohen lab and finished in our lab. | ||
| + | *Overnight cultures of FY4 and FY5 were started. | ||
| + | *An overnight culture of FY4 and FY5 were made in YPD 7 from FY4 plate 7/26/10 and FY5 plate 7/21/10 | ||
| + | <br> | ||
| - | + | ==2010/07/29== | |
| - | + | ||
| - | + | ||
| - | == | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
1. FY4 Cohen T-Mix 0ng YPD Plate Result: A Lawn formed on the plate | 1. FY4 Cohen T-Mix 0ng YPD Plate Result: A Lawn formed on the plate | ||
2. FY4 Cohen T-Mix 0ng G418 Plate Result: Only Dead cells were observed | 2. FY4 Cohen T-Mix 0ng G418 Plate Result: Only Dead cells were observed | ||
| Line 299: | Line 324: | ||
12. FY5 Our T-Mix 500ng G418 Plate Result: 86 colonies were observed, sizes ranged from very small to medium | 12. FY5 Our T-Mix 500ng G418 Plate Result: 86 colonies were observed, sizes ranged from very small to medium | ||
13. Cells with G418 resistance G418 Plate Result: cells appeared to grow in the first 3 lines of the streak, however no colony like units have formed. | 13. Cells with G418 resistance G418 Plate Result: cells appeared to grow in the first 3 lines of the streak, however no colony like units have formed. | ||
| - | Look on 7/30 page for more exact counts and efficiency calculations | + | *Look on 7/30 page for more exact counts and efficiency calculations |
| - | == | + | ==2010/07/30== |
| - | + | ||
1. FY4 Cohen T-Mix 0ng YPD Plate Result: A Lawn formed on the plate | 1. FY4 Cohen T-Mix 0ng YPD Plate Result: A Lawn formed on the plate | ||
2. FY4 Cohen T-Mix 0ng G418 Plate Result: Only Dead cells were observed | 2. FY4 Cohen T-Mix 0ng G418 Plate Result: Only Dead cells were observed | ||
| Line 316: | Line 340: | ||
12. FY5 Our T-Mix 500ng G418 Plate Result: 82 colonies were obsered ranging from small to large efficiency=82cfu/0.5μg=164cfu/μg | 12. FY5 Our T-Mix 500ng G418 Plate Result: 82 colonies were obsered ranging from small to large efficiency=82cfu/0.5μg=164cfu/μg | ||
13. Cells with G418 resistance G418 Plate Result: cells appeared to grow in the first 3 lines of the streak, however no colony like units have formed. | 13. Cells with G418 resistance G418 Plate Result: cells appeared to grow in the first 3 lines of the streak, however no colony like units have formed. | ||
| - | Replated plate 13 from and o/n culture made on 7/29 | + | *Replated plate 13 from and o/n culture made on 7/29 |
| - | + | <br> | |
| - | == | + | ==2010/08/02== |
| - | + | *Growth curve experiments for FY4 and FY5: | |
| - | + | ||
| - | Growth curve experiments for FY4 and FY5: | + | |
Time OD600 of Strian-Fraction Dilution | Time OD600 of Strian-Fraction Dilution | ||
FY4-1 FY4- 1/3 FY4- 1/6 FY4- 1/12 FY5-1 FY5- 1/3 FY5- 1/6 FY5- 1/12 | FY4-1 FY4- 1/3 FY4- 1/6 FY4- 1/12 FY5-1 FY5- 1/3 FY5- 1/6 FY5- 1/12 | ||
| Line 339: | Line 361: | ||
| - | Calculated doubling time for FY4=1:49 | + | *Calculated doubling time for FY4=1:49 |
| - | + | *Calculated doubling time for FY5=2:00 | |
| - | Calculated doubling time for FY5=2:00 | + | <center> |
| - | + | ||
[[Image:WashU_FY4.PNG|400px]] | [[Image:WashU_FY4.PNG|400px]] | ||
| - | |||
| - | |||
| - | |||
[[Image:WashU_FY5.PNG|400px]] | [[Image:WashU_FY5.PNG|400px]] | ||
| + | </center> | ||
| + | <br /> | ||
| - | == | + | ==2010/08/08=== |
| - | Examined The cells transformed on 8/6/2010 | + | *Examined The cells transformed on 8/6/2010 |
| - | Plate | + | Plate |
1. Observed no colonies on the Control | 1. Observed no colonies on the Control | ||
2. Observed a lawn of colonies on 3.4 μg of Linear DNA | 2. Observed a lawn of colonies on 3.4 μg of Linear DNA | ||
| Line 360: | Line 380: | ||
6. Observed a lawn of colonies on 0.8 μg of plasmid 3 hr incubation | 6. Observed a lawn of colonies on 0.8 μg of plasmid 3 hr incubation | ||
7. Observed a lawn of colonies on 0.8 μg of plasmid o/n incubatoin | 7. Observed a lawn of colonies on 0.8 μg of plasmid o/n incubatoin | ||
| - | Four colonies were chosen from the successful linear transformation and streak purified onto a new G418 plate. | + | *Four colonies were chosen from the successful linear transformation and streak purified onto a new G418 plate. |
| - | + | <br> | |
| - | == | + | ==2010/08/10== |
| - | + | *Examined the 4 colonies that were streak purified and saw that none grew. | |
| - | + | *G418 plates were used from the same batch so we weren't sure what the cause of this was. | |
| - | + | *Possibly it is due to the fact a lawn formed. | |
| - | + | *Several colonies at a time were then restreaked from the original successful transformation plate and put onto a new G418 plate | |
| - | == | + | <br> |
| - | + | ==2010/08/16== | |
| - | + | *Yeast Mating: Preformed a yeast mating between the CSxL-FY4 yeast and FY5 yeast according to the following protocol from the clonetech yeast potocol handbook: | |
| - | Yeast Mating: Preformed a yeast mating between the CSxL-FY4 yeast and FY5 yeast according to the following protocol from the clonetech yeast potocol handbook: | + | 2. Yeast mating procedure (standard) |
| - | 2. Yeast mating procedure (standard) | + | a. Pick one colony of each type to use in the mating. Use only large (2–3-mm), fresh (<2-months old) colonies from the working stock plates. |
| - | + | ||
b. Place both colonies in one 1.5-ml microcentrifuge tube containing 0.5 ml of YPD medium. Vortex tubes to completely resuspend the cells. | b. Place both colonies in one 1.5-ml microcentrifuge tube containing 0.5 ml of YPD medium. Vortex tubes to completely resuspend the cells. | ||
| - | + | c. Incubate at 30°C overnight (20–24 hr) with shaking at 200 rpm. | |
d. Spread 100-µl aliquots of the mating culture on the appropriate SD minimal media. | d. Spread 100-µl aliquots of the mating culture on the appropriate SD minimal media. | ||
| - | The following conditions were tested: | + | *The following conditions were tested: |
1. Mating single colony FY5 with single colony FY4-CSxL | 1. Mating single colony FY5 with single colony FY4-CSxL | ||
2. Mating single colony FY5 with a swab of FY4-CsxL colonies | 2. Mating single colony FY5 with a swab of FY4-CsxL colonies | ||
| Line 382: | Line 401: | ||
4. Growth of just a swab of FY4-CSxL colonies | 4. Growth of just a swab of FY4-CSxL colonies | ||
| - | FY4-CSxL Testing: | + | *FY4-CSxL Testing: |
| - | Plated cells on Ura- plates to test of excesision of homologous segment with the following conditions: | + | **Plated cells on Ura- plates to test of excesision of homologous segment with the following conditions: |
1. 100μL of 10:1 dilution of a saturated o/n culture of FY4-CSxL | 1. 100μL of 10:1 dilution of a saturated o/n culture of FY4-CSxL | ||
2. 100μL of 10:1 dilution of a saturated o/n culture of FY4 | 2. 100μL of 10:1 dilution of a saturated o/n culture of FY4 | ||
| - | Second CSxL Transformation: | + | *Second CSxL Transformation: |
| - | Made fresh plates of FY4 and FY5 for a CSxL test transformation. | + | **Made fresh plates of FY4 and FY5 for a CSxL test transformation. |
| - | Amplification of pSB1AT3 | + | *Amplification of pSB1AT3 |
| - | Transformed competent e. coli with DNA from the 2010 iGEM spring distribution wellplate A13 | + | **Transformed competent ''e. coli'' with DNA from the 2010 iGEM spring distribution wellplate A13 |
| + | <br> | ||
| - | == | + | ==2010/08/17== |
| - | Yeast Mating | + | *Yeast Mating |
| - | Plated the o/n culutures form the yeast mating preformed on 8/16 on G418 resistant plates. | + | **Plated the o/n culutures form the yeast mating preformed on 8/16 on G418 resistant plates. |
| - | Yeast Trasnformation on 8/19 | + | *Yeast Trasnformation on 8/19 |
| - | Replated FY4 and FY5 strains because it did not appear that we were going to be able to get single colonies from the cells plated on 8/16. | + | **Replated FY4 and FY5 strains because it did not appear that we were going to be able to get single colonies from the cells plated on 8/16. |
| - | pSB1AT3 Amplificiation | + | *pSB1AT3 Amplificiation |
| - | Approximately 20-30 colonies were observed to have grown on the plate and a single colony was used to create an overnight culture. | + | **Approximately 20-30 colonies were observed to have grown on the plate and a single colony was used to create an overnight culture. |
| + | <br> | ||
| - | == | + | ==2010/08/18== |
| - | Yeast Group | + | *Yeast Group |
| - | Made o/n cultures for 8/19 transformation | + | *Made o/n cultures for 8/19 transformation |
| - | Digested CSxL miniprepped DNA with AVRII and column purified it | + | *Digested CSxL miniprepped DNA with AVRII and column purified it |
| + | <br> | ||
| - | == | + | ==2010/08/19== |
| - | Conducted a Transformation of CSxL using on both the FY4 strain and the FY5 strain | + | *Conducted a Transformation of CSxL using on both the FY4 strain and the FY5 strain |
Time FY4 OD600 FY5 OD600 | Time FY4 OD600 FY5 OD600 | ||
9:00 0.090 0.094 | 9:00 0.090 0.094 | ||
| Line 416: | Line 438: | ||
15:00 0.890 0.910 | 15:00 0.890 0.910 | ||
| - | The Follwoing Conditions were used: | + | *The Follwoing Conditions were used: |
Plate # μg DNA Strain V DNA V T-mix V H20 | Plate # μg DNA Strain V DNA V T-mix V H20 | ||
1 0 FY5 0 345.1 14.9 | 1 0 FY5 0 345.1 14.9 | ||
| Line 424: | Line 446: | ||
5 0.04 FY5 1.49 (1:10) 345.1 13.41 | 5 0.04 FY5 1.49 (1:10) 345.1 13.41 | ||
| - | T-Mix Components: | + | *T-Mix Components: |
| - | 1.92ml 50% PEG | + | 1.92ml 50% PEG |
| - | 288μL 1M LiAc | + | 288μL 1M LiAc |
| - | 80μL Salmon Sperm | + | 80μL Salmon Sperm |
| - | 472.8 μL H20 | + | 472.8 μL H20 |
| - | + | <br> | |
| - | == | + | ==2010/08/20== |
| - | Column purified yesterday's digests | + | *Column purified yesterday's digests |
Name DNA concentration ng/uL | Name DNA concentration ng/uL | ||
1 K58 0.7 | 1 K58 0.7 | ||
| Line 443: | Line 465: | ||
*Note: K = KanMX4, N = NatMX4, P1 = pSB1C3 | *Note: K = KanMX4, N = NatMX4, P1 = pSB1C3 | ||
| - | Ligated digestion products 1-6 into vector | + | *Ligated digestion products 1-6 into vector |
1. 7.5 uL K58 + 1 uL P1 | 1. 7.5 uL K58 + 1 uL P1 | ||
| Line 452: | Line 474: | ||
6. 8 uL N64 + 1 uL P1 | 6. 8 uL N64 + 1 uL P1 | ||
| - | Transformed all ligations and plasmid pSB1C3 with insert BBa_J04450, RFP into E. coli | + | *Transformed all ligations and plasmid pSB1C3 with insert BBa_J04450, RFP into ''E. coli'' |
| - | Plated on LB with chloramphenicol. | + | *Plated on LB with chloramphenicol. |
| - | + | <br> | |
| - | + | ||
| - | == | + | ==2010/09/09== |
| - | Prepared colony PCR of 5 colonies each from linear transformation on 8/5/10 and 8/20/10 | + | *Prepared colony PCR of 5 colonies each from linear transformation on 8/5/10 and 8/20/10 |
| - | Protocol for colony PCR with yeast (from Kim): | + | *Protocol for colony PCR with yeast (from Kim): |
1. Aliquot the amount of water to be used in the PCR reaction into tubes. | 1. Aliquot the amount of water to be used in the PCR reaction into tubes. | ||
2. Take each desired colony and inoculate the appropriate tube. | 2. Take each desired colony and inoculate the appropriate tube. | ||
| Line 467: | Line 488: | ||
5. Proceed with PCR reaction as usual. | 5. Proceed with PCR reaction as usual. | ||
| - | PCR reaction: | + | *PCR reaction: |
5 ul buffer | 5 ul buffer | ||
1 ul DNTP mix | 1 ul DNTP mix | ||
| Line 483: | Line 504: | ||
6. 68 degrees C 10:00 | 6. 68 degrees C 10:00 | ||
7. 4 degrees C forever | 7. 4 degrees C forever | ||
| + | <br> | ||
| - | == | + | ==2010/09/10== |
| - | Gel of colony PCR of Sxl construct showed bands at about 1 kb and no other, showing that the Sxl construct was not integrated into the yeast genome. | + | *Gel of colony PCR of Sxl construct showed bands at about 1 kb and no other, showing that the Sxl construct was not integrated into the yeast genome. |
| - | Sxl was ran on a gel to confirm its length. | + | *Sxl was ran on a gel to confirm its length. |
Latest revision as of 07:15, 27 October 2010

2010/06/24
- plated Yeast Gal-YFP MATa, trp1, leu2::GAL1-YFP URA3/Kanr and Yeast YF4 MATa Prototroph on a Kan plate and a YPD plate
- Started a liquid culture of Yeast YF4 MATa Prototroph in YPD
2010/06/29
- A practice transformation was preformed at the Cohen Lab with Ilaria.
- The OD600 was 0.651 which was low, but used in order to speed up the experiment.
- The PMKV005 loxKanlox Plasmid (McCusker 2005 Yeast, 21: 163-171) was used into transform wildtype yeast.
- The plates used were Kan positive plates created on 6/24.
- However it was later found that these plates might not correctly select for drug resistance.
- Two plates were made, one with the transformant and one with a control.
- The attached procedure was used and annotated during the experiment
2010/07/01
- Examined the Sample and Control plates from the practice transformation on June 29th.
- Two colonies had grown on the Sample plate and none showed on the control plate.
- The low colony number could be due to a low OD600 (0.651) used in the transformation.
- Plated the two colonies on a G418 plate in order to conduct the first round of purification
2010/07/02
- PRACTICE YEAST TRANSFORMATION:
- Colonies grew on the first purification plate for both colony 1 and 2.
- Two colonies were plated from both transformed colony 1 and transformed colony 2 onto a second and final purification plate
- PLATES:
YPD: all yeast grew His-: only 1&2 (WT) yeast grew, (4,6,11,12 did not grow) Kan:3,4,7 grew- 1&2 were selected against but some colonies were visible Ura-: only 1&2 (WT) yeast grew, (11 did not grow)
- Amp: bacteria 8,9,10 grew (other amp plate WT control did not grow)
- Conclusion: all the resistance plates work in that they select against those w/o the resistance; however, there was some low level of growth on some of these plates where there was not supposed to be
2010/07/12
- Streaked out wild type yeast strain FY4 onto a YPD plate and inoculated about 5 mL of YPD- incubate and shake
- Brian: The FY5 freezer stock was accidentally allowed to thaw and therefor a second freezer stock was started by growing an overnight culture inoculated from the original freezer stock.
- The freezer stock was made according to protocol from the cohen lab.
2010/07/13
- Plated FY4 yeast onto a YPD plate to grow up for a practice transformation in case the plate from the freezer stock does not grown since there were no visible growths on it.
- The FY4 was taken from a Ura- plate plated on 6/29 because it had the best colonies out of all the plates in the refrigerator.
- A freezer stock of FY4 was made according to protocol from an overnight culture of the FY4 strain since the original stock was destroyed when plating on 7/12.
2010/07/14
- Yeast colonies were observed on the FY4 plated on 7/12 from the FY4 freezer stock. This plate was brought to the Cohen lab along with liquid YPD-4 and they were used to inoculate a 5ml overnight culture.
2010/07/15
- Yeast Transformation
- Alice and Brian went to the Cohen Lab to preform a yeast transformation.
- Ilaria showed us how to use the nano-drop using the following procedure:
1. Wipe the nanodrop with a kim-wipe 2. Click the shortcut on the computer that is identicle to the logo on the nano drop 3. Choose Nucleic Acid in the program 4. Put on 1 μL of dH2O to clean it 5. Cleantop and bottem with kim wipe 6. Put on 1 μL of blank 7. Clean with Kim wipe 8. Put on 1 μL of plasmid 9. Make sure sample type is set to DNA 50
- Looked at the graph, a spike at 130 is due to salts, a spike at 260 is due to DNA, a spike at 280 is due to protein. *Record the 260/230 ratio and 260/280 ratios, you want then both to be above 1, somewhere between 1-2 is good
- A DNA concentration of ~1ng/ μL was measured which is extremely low, and therefor the plasmid should not be used to preform a yeast transformation.
- This could be a poissible explination for the extremely low transformation efficiency of Transformation-1.
- Because of this it is necessary to amplify the plasmid in E. coli.
- Amplification of PMKV005
- The PMKV005 was to be amplified in E. coli to increase its concentration.
- Vorachek-Warren et. al. 2004 showed that PMKV005 was derived from the pAG26 plasmid which had ampicillin resistance.
- The competent cells were obtained from NEB thawed on ice and transformed with the following protocol.
1. Pipette 1 μL of plasmid into the cells. Flick the tubes very gently; 2. Let them site on ice for 10 minutes; 3. heat shock at 42 degC for 45 seconds; 4. Return tubes on ice; 5. add 250 μL SOC media (LB works too but SC was used); 6. grow cells at 37 degC 225 rpm for 1 hour; Placed in incubator directly to the left of the shaker in the cohen lab 7. Plate 50 μL on a antibiotic resistant plate (which you have warmed up with the lid slightly open for about 20 minutes); 8. incubate overnight at 37 degC.
2010/07/16
- Plated a colony from the PMKV005 e. coli transformation on 7/15 into LB for an overnight culture.
2010/07/17
- Conducted a Miniprep from the o/n culture of PMKV005 transformed E. coli made on 7/16.
- Conducted using the Plasmid DNA Purification Using the QIAprep Spin Miniprep Kit and a Microcentrifuge protcol on page 22 of the attached manual.
- All places when centrifugation is between 30-60 seconds 60 seconds were used.
- The 3 LB plates from the Mol. Bio. group were also examined and no colonies were observed.
2010/07/18
- Made an overnight culture of the FY4 strain using YPD-2
2010/07/19
- Prepared an o/n culture of FY4 strain from plate made on 7/12 for transformation on 7/20
- Nanodropped the PMKV005 plasmid purified on 7/17 and found a concentration of 22.8 μL
- Conducted a Yeast Transformation following the protocol received from Ilaria
- FY4 cells were used from culture started on 7/18.
- Cell inoculated at 8:55am and put in shaker at 37 ° C until 12:30pm when it was transported over to the Cohen lab. OD600 taken at 12:50 measure 0.318. At 2:45 a final OD600 of 0.631 was reached.
- The T-mix was prepared x6 and the salmon sperm was boiled at x9 because some boils off
- 9,10. The cells were transformed with the following mixtures.
Vial # Concentration of DNA (ng/μL) Volume of DNA (μL) Mass of DNA (ng) Volume of T-mix 1 22.8 0.877 20 359 2 22.8 0.439 10 359.5 3 .228 8.77 2 351 4 .228 0.877 0.2 359 5 0 0 0 360
- Our first round of Kan Plates were used to plate the colonies.
2010/07/20
- Attempted to conduct a transformation back at our lab.
- However we were unable to get an OD600 reading.
- The nano-drop gave inconsistent and negligible results.
- Using a spectrophotometer an OD of just 0.004 was recorded, on two separate trials.
- The following procedure was used for the spectrophotometer.
- An o/n culture of the FY4 strain plated on 7/12 was made.
1. Turn on the instrument by rotating the 0%T dial clockwise.
You should hear it click when it turns on.
2. Allow the spec to warm up for at least 15 minutes.
Given the long warm up time we will likely be leaving this device on during periods of heavy use.
3. After the warm-up period, set the wavelength to 600 nm (top knob).
4. Set the filter lever to the appropriate position for the selected wavelength (bottom).
5. Press the MODE button until the display mode is set to FACTOR.
6. Using the INCREASE/DECREASE buttons ensure that the factor is set to 1.0.
7. Press the MODE button until the display mode is set to TRANSMITTANCE.
8. With no cuvette in the holder and the lid closed, turn the 0%T knob until the measured transmittance is 0.0.
This is zeroing the device.
9. Fill a cuvette with 3.0 mL of the medium used to grow the cells (e.g. YPD) and wipe the sides with a KimWipe to eliminate fingerprints and dust particles.
10. Place the cuvette in the compartment.
11. Align the guide mark of the cuvette with the guide mark on the compartment.
12. Close the lid of the compartment.
13. Adjust the TRANSMITTANCE to 100.0 using the 100%T knob (right).
This is essentially the equivalent of when you TARE a balance.
14. Remove the sample.
You may choose to leave this cuvette of YPD for other users as they will likely be performing the same procedure.
Note: the spectrophotometer must be blanked for each user even if a common cuvette is being used.
15. In a new cuvette add 3.0 mL of your cell culture sample.
16. Wipe the cell with a KimWipe and insert into the compartment as in Step 10.
17. Record the value shown in the data display (make sure MODE is ABSORBANCE).
Absorbance is a synonym for OD (optical density).
18. When all measurements are obtained, turn the O%T (left knob) counterclockwise to switch the spec off.
19. Clean your cuvettes thoroughly and dry them completely.
- Taken from http://fg.cns.utexas.edu/fg/course_notebook_chapter_nine.html
2010/07/21
- Measured the OD600 of the overnight FY4 culture using a spectrophotometer and a nanodrop
- Spectrophotometer gave a reading of 1.220
- Nano drop gave readings of .282, 0.347, 0.360
- The nanodrop has a 1mm pathlength while the spectrophotometer has a 1 cm pathlength so there is an expected factor of 10 difference
- We later became aware that the spectrophotometer cannot measure saturated cultures accurately so we conducted a more thorough experiment.
- The culture was diluted 5 fold and the absorbance of both the saturated and diluted cultures were measured.
- Results
Absorbance OD600 Spectrophotomer Saturated Culture 1.055 1.055 Spectrophtometer 1/5 dilution .221 1.105 Nanodrop Saturated Culture Trial 1 0.131 1.31 Nanodrop Saturated Culture Trial 2 0.181 1.81 Nanodrop Saturated Culture Trial 3 0.133 1.33 Nanodrop Saturated Culture Trial 4 0.159 1.59 Nanodrop Saturated Culture Trial 5 0.201 2.01 Nanodrop 1/5 dilution trial 1 0.155 1.55 Nanodrop 1/5 dilution trial 2 2.2 22 Nanodrop 1/5 dilution trial 3 0.712 7.12 Nanodrop 1/5 dilution trial 4 -0.677 -6.77
- From these results we have chosen not to use the nanodrop to measure OD600 of cell cultures because it gives very sporadic readings instead the spectrophotometer will be used.
- An overnight culture of the FY4 strain was started using YPD-1
- The FY5 strain was plated from freezer stock and from the plate with strains 1-12 on it
3. Gal-YFP 4. Gal-CFP 5. Control YFP 6. Control CFP 7. Gal CFP-YFP
2010/07/22
- YPD: The YPD made in the cohen lab on 7/19 was divided up into separate bottles
- Transformation: A culture was started using 50 ml of the remainder of the YPD from the first batch and 750 μL of overnight culture.
- An OD600 reading was taken 6 hours later and it recorded only .8
- Therefore the transformation was aborted due to slow growth.
- Growth Curve Analysis: A 50ml culture was started with an OD600 of 0.10 in a 250 ml flask, capped with a medal top and place on the shaker at 100rpm in 30 degC.
- OD600 readings were taken every hour.
- Overnight culture OD600: 5X dilution OD600=0.392 therefor the OD600 of the overnight culture is 1.69
- 1.96* culture volume/50ml=0.1 therefor the culture volume used was 2.55ml
Time Time Passed OD600 Notes 9:40:00 0 0.113 10:40:00 1:00:00 0.14 11:40:00 2:00:00 0.18 12:42:00 3:02:00 0.252 13:46:00 4:06:00 0.354 14:40:00 5:00:00 0.485 15:39:00 5:59:00 0.642 16:40:00 7:00:00 0.845 17:37:00 7:57:00 1.356 Used 3x dilution 18:39:00 8:59:00 1.938 Used 3x dilution 19:10:00 9:30:00 2.136 Used 3x dilution 23:02:00 13:22:00 4.536 used 6x diltution
2010/07/23
- Contamination Detection
- The overnight culture created on 7/23 was examined under a light microscope for contamination.
- 100 μL of o/n culture was placed in a microcentrifuge tube and then vortexed to seperate the cells.
- 10 μL of the culture from the microcentrifuge tube was then placed on a glass slide and covered with a coverslip.
- The cells were observed at 40x with a Nikon Eclipse 200 microscope.
- Small circular cells were observed moving across the screen at a slow pace. The cells formed groupings of 2-3.
- Nothing other of note was observed.
- Conclusions:
- It was concluded that no observable contamination of the cell culture was seen.
- Yeast Transformation with pMKV005: attempt #2
- Overnight culture created on 7/22 was brought to an OD of 0.174 to grow for around 5 hours to a final OD of 0.7.
- At this time, the yeast was transformed with 20 ng, 2ng, and 0.2 ng of pMKV005 and plated on YPD/Kan plates.
- A control was also made. No colonies were seen on any of the plates after 2 days of incubation.
2010/07/26
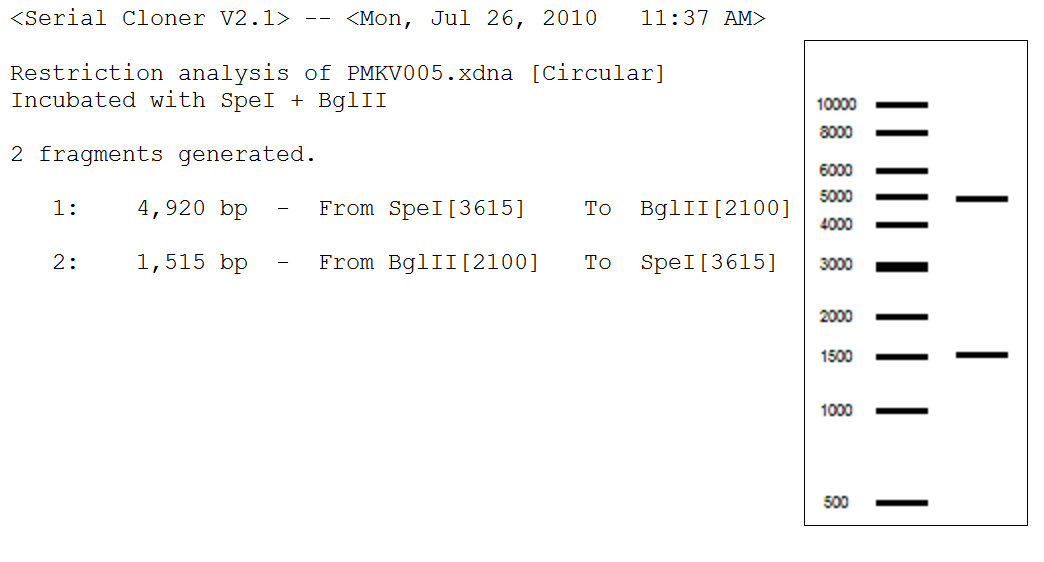
- Ran a digest and gel with the mol. bio. group. (All in μL)
1 2 3 4 5 6 7
Vector PMKV005 PMKV005 PMKV005 Ksa1 Ksa2 Ksb1 Ksb2 Water 22.5 32.5 33.5 39.5 39.5 39.5 39.5 DNA 20 10 10 3 3 3 3 Buffer 2 5 5 5 5 5 5 5 Enzyme 1 SpeI SpeI BglII EcoRI EcoRI EcoRI EcoRI Enzyme 2 BglII BglII - BamHI BamHI BamHI BamHI BSA 0.5 0.5 0.5 0.5 0.5 0.5 0.5
- Expected results of PMKV005 digest:
2010/07/27
- Replated the FY4 plate from 7/26 since the colonies were too dense.
- Two minipreps were preformed on 2 different cultures that had been transformed with the PMKV005 Plasmid. The Qiagen protocol was followed except when the cells were initially spun down the centrifuge was set to 3000 rpm for 10 minutes. To increase the concentration of one of the samples a 30 μL elution instead of a 50 μL elution was used.
- The samples were nanodroped following standard protocol and the follow data was obtained:
1: 50 μL elution 262.4 ng/μL 260/280=1.89 260/230=2.39 2: 30μL elution 402.5 ng/μL 260/280=1.88 260/230=2.30
- Preformed a Yeast transformation follwing illaria's procedure at our lab.
- The following conditions were used
1. FY4 Cohen T-Mix 0ng YPD Plate 2. FY4 Cohen T-Mix 0ng G418 Plate 3. FY4 Cohen T-mix 100ng G418 Plate 4. FY4 Cohen T-Mix 500ng G418 Plate 5. /NOT USED/FY4 Our T-mix 100ng G418 Plate 6. FY4 Our T-Mix 500ng G418 Plate 7. FY5 Cohen T-Mix 0ng YPD Plate 8. FY5 Cohen T-Mix 0ng G418 Plate 9. FY5 Cohen T-Mix 100ng G418 Plate 10. FY5 Cohen T-Mix 500ng G418 Plate 11. /NOT USED/FY5 Our T-mix 100ng G418 Plate 12. FY5 Our T-Mix 500ng G418 Plate 13. Cells with G418 resistance G418 Plate
- The experiment was designed to test the following conditions:
- Test whether the cells are dieing during the transformation
1. FY4 Cohen T-Mix 0ng YPD Plate 7. FY5 Cohen T-Mix 0ng YPD Plate
- Test the control to insure transformation is necessary for colony growth
2. FY4 Cohen T-Mix 0ng G418 Plate 8. FY5 Cohen T-Mix 0ng G418 Plate
- Test whether a G418 Strain can survive on our G418 Plates
13. Cells with G418 resistance G418 Plate
- Test the affect of using 500 ng of DNA in place of 100ng of DNA
3. FY4 Cohen T-mix 100ng G418 Plate 4. FY4 Cohen T-Mix 500ng G418 Plate 9. FY5 Cohen T-Mix 100ng G418 Plate 10. FY5 Cohen T-Mix 500ng G418 Plate
- Experimental Notes
- OD600 Readings
Time OD600
Overnight FY4 0.454X6=2.724* Overnight FY5 0.686x65=4.116* FY4 9:10am 0.101* FY5 9:10am 0.082* FY4 12:20pm 0.200 FY5 12:20pm 0.224 FY4 3:15 pm 0.564 FY5 3:15pm 0.825 FY4 4:05pm 0.734 FY5 4:05pm 1.110 FY4 4:40pm 0.338x3= 1.014 FY5 4:40pm 0.606x3=1.818
- The first 4 readins were actually taken at wavelength 579nm but this should have very small affect on the values
- During the heat bath step it was observed that the cells settled at the bottem of the microcentrifuge tubes, we are not sure if this is normal or what affect it would have on the experiment.
2010/07/28
- YPD and G418 plates were started at the cohen lab and finished in our lab.
- Overnight cultures of FY4 and FY5 were started.
- An overnight culture of FY4 and FY5 were made in YPD 7 from FY4 plate 7/26/10 and FY5 plate 7/21/10
2010/07/29
1. FY4 Cohen T-Mix 0ng YPD Plate Result: A Lawn formed on the plate 2. FY4 Cohen T-Mix 0ng G418 Plate Result: Only Dead cells were observed 3. FY4 Cohen T-mix 100ng G418 Plate Result: 8 colonies were observed, all of them were small in size 4. FY4 Cohen T-Mix 500ng G418 Plate Result:4 colonies were obsered, all of them were small in size 5. /NOT USED/ 6. FY4 Our T-Mix 500ng G418 Plate Result: No colonies were observed 7. FY5 Cohen T-Mix 0ng YPD Plate Result: A Lawn formed 8. FY5 Cohen T-Mix 0ng G418 Plate Result: No colonies were observed 9. FY5 Cohen T-Mix 100ng G418 Plate Result: No colonies were observed 10. FY5 Cohen T-Mix 500ng G418 Plate Result: 50 colonies were observed, sizes ranged from very small to medium 11. /NOT USED/ 12. FY5 Our T-Mix 500ng G418 Plate Result: 86 colonies were observed, sizes ranged from very small to medium 13. Cells with G418 resistance G418 Plate Result: cells appeared to grow in the first 3 lines of the streak, however no colony like units have formed.
- Look on 7/30 page for more exact counts and efficiency calculations
2010/07/30
1. FY4 Cohen T-Mix 0ng YPD Plate Result: A Lawn formed on the plate 2. FY4 Cohen T-Mix 0ng G418 Plate Result: Only Dead cells were observed 3. FY4 Cohen T-mix 100ng G418 Plate Result: 14 colonies were observed, ranged from small to medium Efficiency=14colonies/0.1μg=140cfu/μg 4. FY4 Cohen T-Mix 500ng G418 Plate Result: 8 colonies were obsered, all of them were small in size Efficiency=8 colonies/0.5μg=16cfu/μg 5. /NOT USED/ 6. FY4 Our T-Mix 500ng G418 Plate Result: 6 colonies were observed, all of them small in size Efficiency=6colonies/0.5μg=12 cfu/μg 7. FY5 Cohen T-Mix 0ng YPD Plate Result: A Lawn formed 8. FY5 Cohen T-Mix 0ng G418 Plate Result: No colonies were observed 9. FY5 Cohen T-Mix 100ng G418 Plate Result: No colonies were observed efficiency =0cfu/μg 10. FY5 Cohen T-Mix 500ng G418 Plate Result: 32 colonies were observed ranging from medium to large Efficiency: 32/0.5μg=64cfu/μg 11. /NOT USED/ 12. FY5 Our T-Mix 500ng G418 Plate Result: 82 colonies were obsered ranging from small to large efficiency=82cfu/0.5μg=164cfu/μg 13. Cells with G418 resistance G418 Plate Result: cells appeared to grow in the first 3 lines of the streak, however no colony like units have formed.
- Replated plate 13 from and o/n culture made on 7/29
2010/08/02
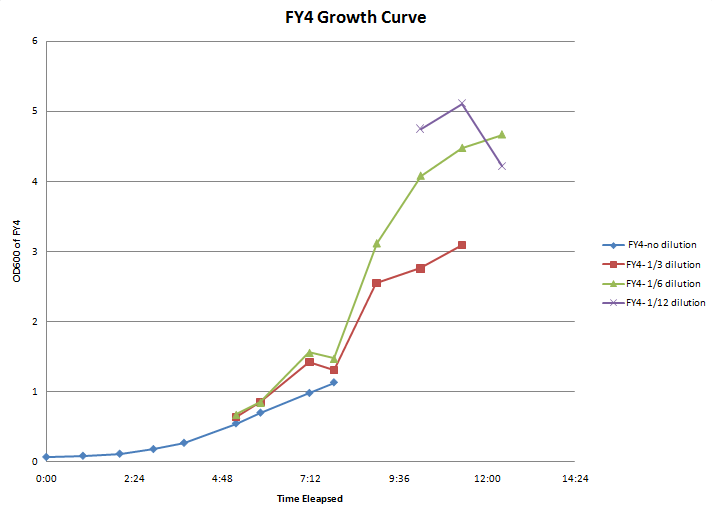
- Growth curve experiments for FY4 and FY5:
Time OD600 of Strian-Fraction Dilution FY4-1 FY4- 1/3 FY4- 1/6 FY4- 1/12 FY5-1 FY5- 1/3 FY5- 1/6 FY5- 1/12 9:10 0.071 0.062 10:10 0.088 0.068 11:10 0.117 0.086 12:05 0.183 0.121 12:55 0.272 0.136 2:20 0.548 0.215 0.113 0.285 3:00 0.700 0.285 0.142 0.410 4:20 0.980 0.476 0.260 0.484 5:00 1.13 0.436 0.246 0.326 0.176 6:10 0.850 0.520 0.520 0.275 7:22 0.920 0.680 0.396 0.774 0.488 8:30 1.030 0.746 0.426 0.965 0.588 0.363 9:35 0.778 0.352 0.614 0.350
- Calculated doubling time for FY4=1:49
- Calculated doubling time for FY5=2:00
2010/08/08=
- Examined The cells transformed on 8/6/2010
Plate 1. Observed no colonies on the Control 2. Observed a lawn of colonies on 3.4 μg of Linear DNA 3. Observed a lawn of colonies on 1.6 μg of plasmid 3hr incubation 4. Observed a lawn of colonies on 1.2 μg of plasmid 3hr incubation 5. Plate not used 6. Observed a lawn of colonies on 0.8 μg of plasmid 3 hr incubation 7. Observed a lawn of colonies on 0.8 μg of plasmid o/n incubatoin
- Four colonies were chosen from the successful linear transformation and streak purified onto a new G418 plate.
2010/08/10
- Examined the 4 colonies that were streak purified and saw that none grew.
- G418 plates were used from the same batch so we weren't sure what the cause of this was.
- Possibly it is due to the fact a lawn formed.
- Several colonies at a time were then restreaked from the original successful transformation plate and put onto a new G418 plate
2010/08/16
- Yeast Mating: Preformed a yeast mating between the CSxL-FY4 yeast and FY5 yeast according to the following protocol from the clonetech yeast potocol handbook:
2. Yeast mating procedure (standard) a. Pick one colony of each type to use in the mating. Use only large (2–3-mm), fresh (<2-months old) colonies from the working stock plates. b. Place both colonies in one 1.5-ml microcentrifuge tube containing 0.5 ml of YPD medium. Vortex tubes to completely resuspend the cells. c. Incubate at 30°C overnight (20–24 hr) with shaking at 200 rpm. d. Spread 100-µl aliquots of the mating culture on the appropriate SD minimal media.
- The following conditions were tested:
1. Mating single colony FY5 with single colony FY4-CSxL 2. Mating single colony FY5 with a swab of FY4-CsxL colonies 3. Growth of just a single FY4-CSxL colony 4. Growth of just a swab of FY4-CSxL colonies
- FY4-CSxL Testing:
- Plated cells on Ura- plates to test of excesision of homologous segment with the following conditions:
1. 100μL of 10:1 dilution of a saturated o/n culture of FY4-CSxL 2. 100μL of 10:1 dilution of a saturated o/n culture of FY4
- Second CSxL Transformation:
- Made fresh plates of FY4 and FY5 for a CSxL test transformation.
- Amplification of pSB1AT3
- Transformed competent e. coli with DNA from the 2010 iGEM spring distribution wellplate A13
2010/08/17
- Yeast Mating
- Plated the o/n culutures form the yeast mating preformed on 8/16 on G418 resistant plates.
- Yeast Trasnformation on 8/19
- Replated FY4 and FY5 strains because it did not appear that we were going to be able to get single colonies from the cells plated on 8/16.
- pSB1AT3 Amplificiation
- Approximately 20-30 colonies were observed to have grown on the plate and a single colony was used to create an overnight culture.
2010/08/18
- Yeast Group
- Made o/n cultures for 8/19 transformation
- Digested CSxL miniprepped DNA with AVRII and column purified it
2010/08/19
- Conducted a Transformation of CSxL using on both the FY4 strain and the FY5 strain
Time FY4 OD600 FY5 OD600 9:00 0.090 0.094 13:10 0.400 0.422 14:00 0.604 0.626 15:00 0.890 0.910
- The Follwoing Conditions were used:
Plate # μg DNA Strain V DNA V T-mix V H20 1 0 FY5 0 345.1 14.9 2 4 FY4 14.9 345.1 0 3 4 FY5 14.9 345.1 0 4 0.4 FY5 1.49 345.1 13.41 5 0.04 FY5 1.49 (1:10) 345.1 13.41
- T-Mix Components:
1.92ml 50% PEG 288μL 1M LiAc 80μL Salmon Sperm 472.8 μL H20
2010/08/20
- Column purified yesterday's digests
Name DNA concentration ng/uL 1 K58 0.7 2 N58 1.2 3 K60 1.8 4 N60 1.6 5 K64 2.2 6 N64 1.3 7 P1 3.6
- Note: K = KanMX4, N = NatMX4, P1 = pSB1C3
- Ligated digestion products 1-6 into vector
1. 7.5 uL K58 + 1 uL P1 2. 8 uL N58 + 0.5 uL P1 3. 7.5 uL K60 + 1 uL P1 4. 7.5 uL N60 + 1 uL P1 5. 7.5 uL K64 + 1 uL P1 6. 8 uL N64 + 1 uL P1
- Transformed all ligations and plasmid pSB1C3 with insert BBa_J04450, RFP into E. coli
- Plated on LB with chloramphenicol.
2010/09/09
- Prepared colony PCR of 5 colonies each from linear transformation on 8/5/10 and 8/20/10
- Protocol for colony PCR with yeast (from Kim):
1. Aliquot the amount of water to be used in the PCR reaction into tubes. 2. Take each desired colony and inoculate the appropriate tube. 3. Freeze the colonies in the -80 degree Celsius freezer for 10 minutes. 4. Boil the colonies at 99 degrees Celsius for 5 minutes. 5. Proceed with PCR reaction as usual.
- PCR reaction:
5 ul buffer 1 ul DNTP mix 2.5 ul 1x p32 2.5 ul 1x p33 0.5 ul Taq 44 ul water 55.5 ul reaction
1. 94 degrees C 2:00 2. 94 degrees C 0:30 3. 65 degrees C 1:00 4. 68 degrees C 2:30 5. 2x35 6. 68 degrees C 10:00 7. 4 degrees C forever
2010/09/10
- Gel of colony PCR of Sxl construct showed bands at about 1 kb and no other, showing that the Sxl construct was not integrated into the yeast genome.
- Sxl was ran on a gel to confirm its length.
 "
"