Team:UNIPV-Pavia/Calendar/October/settimana1
From 2010.igem.org
m (→October, 3rd) |
m (→October, 10th) |
||
| (58 intermediate revisions not shown) | |||
| Line 32: | Line 32: | ||
<html><a name="indice"/></html> | <html><a name="indice"/></html> | ||
| + | ==October, 4th== | ||
| + | ON PCR run and than gel run. | ||
| + | [[Image:UNIPV10_04_10_10_I74_massive_PCR.jpg|thumb|200px|center|I74 massive PCR screening.]] | ||
| - | + | As you can see we had the same results as previous PCR. We decided to digest I74-7/8 (two different genotypes) but during centrifugation step for mini we discovered a red pellet in the first one; so we threw it away. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | |||
Miniprep and quantification of: | Miniprep and quantification of: | ||
| - | * | + | *INTEIN_C3: 69 ng/ul |
| - | * | + | *I74-8: 78,2 ng/ul |
| + | *I54: 130,9 ng/ul | ||
| + | *I35: 69 ng/ul | ||
| - | + | Digest: | |
| + | *INTEIN_C3: E-X | ||
| + | *I74-8: E-S (it was also a screening after not clear PCR results) | ||
| + | *I54: E-X | ||
| + | *I35: E-P | ||
| - | + | Gel run/cut/quantification: | |
| + | [[Image:UNIPV10_04_10_10_phasins_digest.jpg|thumb|200px|center|Digest.]] | ||
| + | I54 wasn't in gel. ??? | ||
| - | + | *INTEIN_C3 (E-X): | |
| + | *I35 (E-P): | ||
| + | *I74-8 (E-S) | ||
| + | |||
| + | Ligations: | ||
| + | *I78: I74 (E-S) + INTEIN_C3 (E-X) | ||
| - | + | Transormation (1ul) into ''E. coli'' TOP10 of: | |
| - | + | *I75 | |
| - | + | *I77 | |
| + | They were plated on LB+Amp agar plates and let grow ON at +37°C, 220 rpm. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
---- | ---- | ||
| - | |||
| - | |||
| - | == | + | <font color='red' size='+2'>We are now moving all our parts to <partinfo>pSB1C3</partinfo> plasmid</font> |
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==October, 5th== | ==October, 5th== | ||
| + | |||
| + | I75 and I77 plates showed lots of red colonies. It's unexplainable, nothing should have carried RFPs or something similiar. Probably we had a kind of contamination during digestion or ligation. So we decided to perform digestions again: | ||
| + | *I55: E-S | ||
| + | *I37: E-X | ||
| + | *I57: E-X | ||
| + | *I31: E-S | ||
| + | and we added | ||
| + | *I54: (E-X) | ||
| + | Digestions were let at +37°C ON. | ||
| + | |||
| + | ---- | ||
| + | Transformation (1ul) of | ||
| + | *I78: I74 (E-S) + INTEIN_C3 (E-X) | ||
| + | into ''E. coli'' TOP10. In order to check our ''E. coli'' TOP10 competent cells we plated them on LB+Amp and Cm agar plates to see if something grew. | ||
| + | |||
| + | ---- | ||
| + | ON ligation of: | ||
| + | *I79: I60 (E-S) + INTEIN_C3 (E-X) | ||
| + | |||
| + | ---- | ||
| + | <font color='red' size='+2'>We are now moving all our parts to <partinfo>pSB1C3</partinfo> plasmid</font> | ||
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==October, 6th== | ==October, 6th== | ||
| + | ''E. coli'' TOP10 plated on LB+Amp agar plate showed red colonies!!! We will perform competentization again ASAP (competent cells were conteminated, not DNA !!!). | ||
| + | |||
| + | ---- | ||
| + | Transformation of available ligations I75 and I77 (1ul) into ''E. coli'' TOP10, I79 into ''E. coli'' STBL3. They were plated on LB+Amp agar plates and let grow ON at +37°C, 220 rpm. | ||
| + | |||
| + | ---- | ||
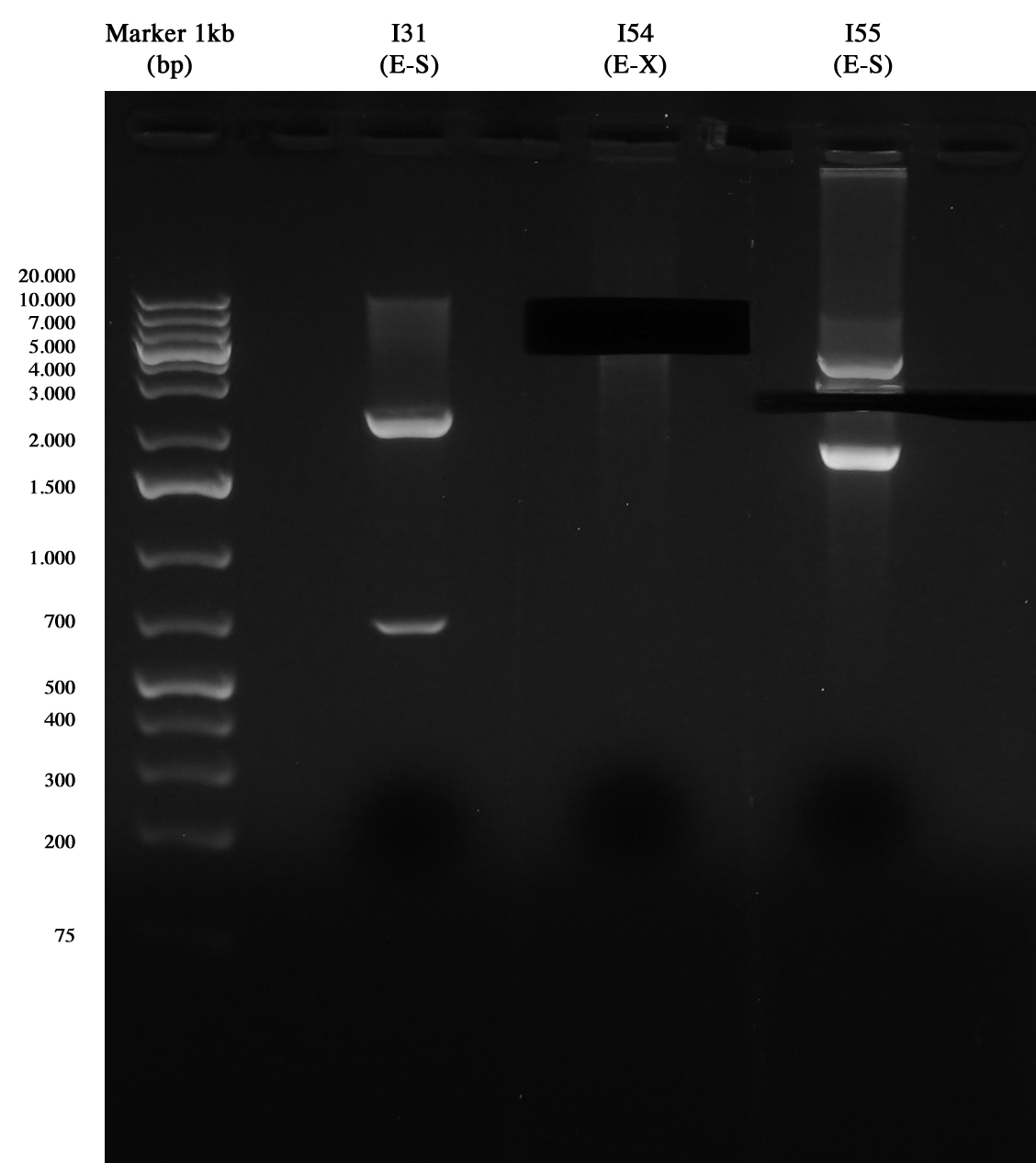
| + | Gel run and cut for ON digestions: | ||
| + | [[Image:UNIPV10_07_10_10_digestioni_giacomo_gel_piccolo.jpg|thumb|200px|center|Digest (I31, I37, I57 weren't necessary anymore).]] | ||
| + | and Nanodrop quantification: | ||
| + | *I54 (E-X): 26 ng/ul | ||
| + | *I55 (E-S): 10 ng/ul | ||
| + | |||
| + | We finally performed ON ligation | ||
| + | *I76: I55 (E-S) + I54 (E-X) | ||
| + | |||
| + | ---- | ||
| + | Colony PCR screening for I78 (eleven colonies were picked from LB+Cm agar plate and were inoculated into 5 ml LB+Cm34 and let grow ON for stock). | ||
| + | PCR was performed this day but it would have been run the following one (too late this evening). | ||
| + | |||
| + | |||
| + | ---- | ||
| + | <font color='red' size='+2'>We are now moving all our parts to <partinfo>pSB1C3</partinfo> plasmid</font> | ||
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==October, 7th== | ==October, 7th== | ||
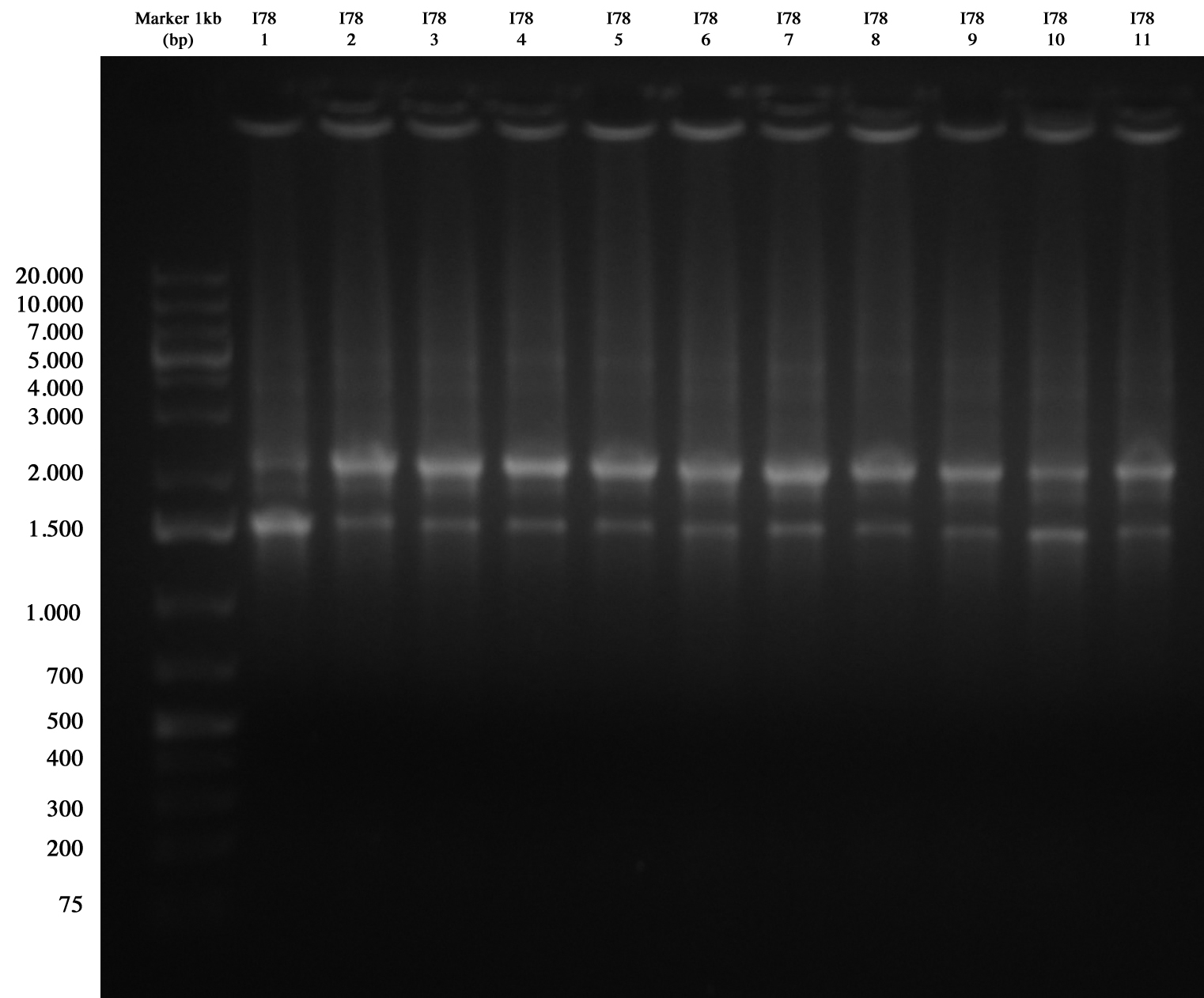
| + | I78 screening gel run | ||
| + | [[Image:UNIPV10_07_10_10_PCR_I78.jpg|thumb|200px|center|Screening PCR for I78 (we cat blank from this picture but it was very clear, trust me).]] | ||
| + | We had extraband in all samples (similar result in I74 PCR: after digest we had a confirmation that it was right. Probably here it was the same problem: very similar repeated sequences give strange PCR; however we sent it sequencing). | ||
| + | |||
| + | Glycerol stock and miniprep for | ||
| + | *I78-3: 153,8 ng/ul | ||
| + | |||
| + | ---- | ||
| + | Colony PCR for screening of I79 (eleven colonies were picked). | ||
| + | [[Image:UNIPV10_07_10_10_PCR_I79.jpg|thumb|200px|center|Screening PCR for I79.]] | ||
| + | |||
| + | Same as the previous PCR. We took I79-2 that was inoculated into LB+Cm34 and let grow ON at +37°C, 220 rpm. | ||
| + | ---- | ||
| + | Transformation (1ul) of | ||
| + | *I0_1C3 | ||
| + | *I3_1C3 | ||
| + | *I7_1C3 | ||
| + | *I8_1C3 | ||
| + | *I9_1C3 | ||
| + | *I12_1C3 | ||
| + | *I76 | ||
| + | into ''E. coli'' DH5-alpha. | ||
| + | |||
| + | They were plated on LB+Cm34 agar plates (except for I76 that was plated on LB+Amp agar plate) and let grow ON at +37°C. | ||
| + | |||
| + | ---- | ||
| + | <font color='red' size='+2'>We are now moving all our parts to <partinfo>pSB1C3</partinfo> plasmid</font> | ||
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==October, 8th== | ==October, 8th== | ||
| + | I76 agar plate showed two/three colonies. It was stored at +4°C; colonies would have been screened the following week. | ||
| + | |||
| + | ---- | ||
| + | |||
| + | Glycerol stock, miniprep and Nanodrop quantification of: | ||
| + | *I79-2: 95,3 ng/ul | ||
| + | |||
| + | This DNA (and that from I55 and I78 too) was sent to BMR Genomics for sequencing. | ||
| + | ---- | ||
| + | <font color='red' size='+2'>We are now moving all our parts to <partinfo>pSB1C3</partinfo> plasmid</font> | ||
| + | |||
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==October, 9th== | ==October, 9th== | ||
| + | |||
| + | <font color='red' size='+2'>We are now moving all our parts to <partinfo>pSB1C3</partinfo> plasmid</font> | ||
| + | ---- | ||
| + | |||
| + | A new part was cloned with these ones, named I80. It is the basic part to build self-inducible promoters and was obtained with the following ligation: | ||
| + | |||
| + | I80=I3(in <partinfo>pSB1C3</partinfo>) (S-P) + <partinfo>BBa_F2620</partinfo> (X-P) | ||
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
| + | |||
| + | ==October, 10th== | ||
| + | Inoculum into 4 ml LB+Amp of: | ||
| + | *I75-1/2/3 | ||
| + | *I76-1/2 | ||
| + | *I77-1/2/3 | ||
| + | for E-P screening; | ||
| + | *I47 | ||
| + | *I48 | ||
| + | *I49 | ||
| + | for transfer into <partinfo>pSB1C3</partinfo>. | ||
| + | |||
| + | They were let grow at +37°C, 220 rpm. | ||
| + | |||
| + | ---- | ||
| + | Transformation of I80 ligation. It was plated on LB+Cm34 agar plate and let grow ON at +37°C. | ||
| + | |||
| + | ---- | ||
| + | <font color='red' size='+2'>We are now moving all our parts to <partinfo>pSB1C3</partinfo> plasmid</font> | ||
| + | |||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
| + | |||
<!-- table previous next week --> | <!-- table previous next week --> | ||
| Line 102: | Line 214: | ||
<!-- fine table previous next week --> | <!-- fine table previous next week --> | ||
</td> | </td> | ||
| + | |||
| + | |||
| + | |||
<td width="15%" align="right" valign="top"> | <td width="15%" align="right" valign="top"> | ||
Latest revision as of 16:38, 23 October 2010
|
|
|||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||
 "
"