Team:UCL London/Week 5
From 2010.igem.org
m |
|||
| (13 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
__NOTOC__ | __NOTOC__ | ||
=Preparation week - Week 5= | =Preparation week - Week 5= | ||
| - | + | '''''26th July - 1st August''''' | |
| - | + | [[Image:UCL-Plate1.JPG|300px|right]] | |
We started transformation of the DNA we need from the iGEM registry plates today. WE had 11 parts to transform, so Xiang and the team had quit a tough few days ahead of them. | We started transformation of the DNA we need from the iGEM registry plates today. WE had 11 parts to transform, so Xiang and the team had quit a tough few days ahead of them. | ||
We proceeded by carrying out a transformation Transformation from the iGEM plates of the parts required, 11 to be precise, with the results expected the following morning | We proceeded by carrying out a transformation Transformation from the iGEM plates of the parts required, 11 to be precise, with the results expected the following morning | ||
| - | |||
| - | |||
| - | |||
| - | |||
{{:Team:UCL_London/clear}} | {{:Team:UCL_London/clear}} | ||
| - | + | [[Image:UCL-Plate2.JPG|300px|left]] | |
| - | + | ||
The results of the transformation on the previous day were confirmedd today...the e.coli cells clearly are not fond of us. Plate 9 (pSB1T3) had no growth on agar plate, and also ontrol plate for chloramphenicol had growth. | The results of the transformation on the previous day were confirmedd today...the e.coli cells clearly are not fond of us. Plate 9 (pSB1T3) had no growth on agar plate, and also ontrol plate for chloramphenicol had growth. | ||
| Line 22: | Line 17: | ||
• Make new 1000X chloramphenicol stock. | • Make new 1000X chloramphenicol stock. | ||
• Transformation of part 15,16,17. | • Transformation of part 15,16,17. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
{{:Team:UCL_London/clear}} | {{:Team:UCL_London/clear}} | ||
| - | |||
[[Image:UCL-Gel.jpg|350px|right]] | [[Image:UCL-Gel.jpg|350px|right]] | ||
| Line 50: | Line 27: | ||
• Run a diagnostic gel on obtained DNA | • Run a diagnostic gel on obtained DNA | ||
• Repick 2 (C-G), 5(C-G), 6(C-G), 8(C-G), 11(C-G), together with plate 15(A&B) & 17(A&B). | • Repick 2 (C-G), 5(C-G), 6(C-G), 8(C-G), 11(C-G), together with plate 15(A&B) & 17(A&B). | ||
| - | |||
| - | |||
| - | |||
| - | |||
| Line 59: | Line 32: | ||
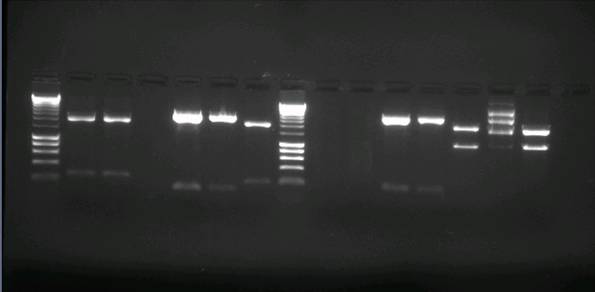
Load from left to right as follows; HyperLadder I was used as marker. | Load from left to right as follows; HyperLadder I was used as marker. | ||
L 1A 1B 2B 3A 3B 4B L 6A 6B 7A 7B 8B 10A 10B | L 1A 1B 2B 3A 3B 4B L 6A 6B 7A 7B 8B 10A 10B | ||
| + | |||
In summary, 2B & 4B had no DNA. 10A contained wrong plasmid or genomic DNA. | In summary, 2B & 4B had no DNA. 10A contained wrong plasmid or genomic DNA. | ||
| Line 64: | Line 38: | ||
Clearly, it looked like it'll take some time for us to get fully acquanted with the e.coli cells, Results 2-6 had no growth in 2ml LB. Maybe if we called ouirselves "don colioness" they might start showing some respect, maybe,....we'll see. | Clearly, it looked like it'll take some time for us to get fully acquanted with the e.coli cells, Results 2-6 had no growth in 2ml LB. Maybe if we called ouirselves "don colioness" they might start showing some respect, maybe,....we'll see. | ||
| - | |||
{{:Team:UCL_London/clear}} | {{:Team:UCL_London/clear}} | ||
| - | + | [[Image:UCL-meetingportico.JPG|500px|thumb|left]] | |
| - | + | ||
| - | [[Image:UCL-meetingportico.JPG| | + | |
This Thursday meeting started as a one time event..and became like a bad habit..but in a good way. The portico seams to provide a very inspiring atmosphere for the iGEM UCL members to discuss their weekly progress and the tasks assigned to each one. For once more, a lot of ideas regarding the project were circulated. | This Thursday meeting started as a one time event..and became like a bad habit..but in a good way. The portico seams to provide a very inspiring atmosphere for the iGEM UCL members to discuss their weekly progress and the tasks assigned to each one. For once more, a lot of ideas regarding the project were circulated. | ||
| Line 79: | Line 50: | ||
{{:Team:UCL_London/clear}} | {{:Team:UCL_London/clear}} | ||
| - | + | [[Image:UCL-Xianglab.jpg|400px|right]] | |
| - | + | ||
Ahaa, the don coliones it is....the results of yesterdays experiments were out; | Ahaa, the don coliones it is....the results of yesterdays experiments were out; | ||
| Line 117: | Line 87: | ||
{{:Team:UCL_London/clear}} | {{:Team:UCL_London/clear}} | ||
| - | |||
| - | + | [https://2010.igem.org/Team:UCL_London/Week_6 '''Week 6'''] | |
| - | + | ||
| - | + | ||
| + | [https://2010.igem.org/Team:UCL_London/Week_1 '''1'''] [https://2010.igem.org/Team:UCL_London/Week_2 '''2'''] [https://2010.igem.org/Team:UCL_London/Week_3 '''3'''] [https://2010.igem.org/Team:UCL_London/Week_4 '''4'''] [https://2010.igem.org/Team:UCL_London/Week_5 '''5'''] [https://2010.igem.org/Team:UCL_London/Week_6 '''6'''] [https://2010.igem.org/Team:UCL_London/Week_7a '''7'''] [https://2010.igem.org/Team:UCL_London/Week_8 '''8'''] [https://2010.igem.org/Team:UCL_London/Week_9 '''9'''] [https://2010.igem.org/Team:UCL_London/Week_10 '''10'''] [https://2010.igem.org/Team:UCL_London/Week_11 '''11'''] [https://2010.igem.org/Team:UCL_London/Week_12 '''12'''] [https://2010.igem.org/Team:UCL_London/Week_13 '''13'''] [https://2010.igem.org/Team:UCL_London/Week_14 '''14'''] | ||
{{:Team:UCL_London/templates/v2/footerFullWidth}} | {{:Team:UCL_London/templates/v2/footerFullWidth}} | ||
Latest revision as of 20:09, 26 October 2010
Preparation week - Week 5
26th July - 1st August
We started transformation of the DNA we need from the iGEM registry plates today. WE had 11 parts to transform, so Xiang and the team had quit a tough few days ahead of them. We proceeded by carrying out a transformation Transformation from the iGEM plates of the parts required, 11 to be precise, with the results expected the following morning
The results of the transformation on the previous day were confirmedd today...the e.coli cells clearly are not fond of us. Plate 9 (pSB1T3) had no growth on agar plate, and also ontrol plate for chloramphenicol had growth.
• Pick 2 different colonies from each plate; inoculate in 2ml LB+antibiotics overnight (O/N). • Make new 1000X chloramphenicol stock. • Transformation of part 15,16,17.
• Miniprep & nanodrop (to measure DNA concentration)
• Run a diagnostic gel on obtained DNA
• Repick 2 (C-G), 5(C-G), 6(C-G), 8(C-G), 11(C-G), together with plate 15(A&B) & 17(A&B).
Gel Result:
Load from left to right as follows; HyperLadder I was used as marker.
L 1A 1B 2B 3A 3B 4B L 6A 6B 7A 7B 8B 10A 10B
In summary, 2B & 4B had no DNA. 10A contained wrong plasmid or genomic DNA.
Results on next day: 2-6 had no growth in 2ml LB.
Clearly, it looked like it'll take some time for us to get fully acquanted with the e.coli cells, Results 2-6 had no growth in 2ml LB. Maybe if we called ouirselves "don colioness" they might start showing some respect, maybe,....we'll see.
This Thursday meeting started as a one time event..and became like a bad habit..but in a good way. The portico seams to provide a very inspiring atmosphere for the iGEM UCL members to discuss their weekly progress and the tasks assigned to each one. For once more, a lot of ideas regarding the project were circulated.
Miniprep of 8DEF, 15AB, 17AB, & 11D. Transformation of part 13, 14, 18-23. Pick colonies from plate 2, 5, 6; grow in LB and TB.
Ahaa, the don coliones it is....the results of yesterdays experiments were out;
2C (TB), 5E-5G (TB), 6E-6G(TB), 6D (LB) --- growth. Plates:
13 Plate √ 14 Plate √ 18 Plate √ 19 X 20 Plate √? (~ 5 dark colonies) 21 X 22 X 23 X
we have the following parts (confirmed by gel electrophoresis):
Inducible promoter lacI
Possible hypoxia promoter from UCL iGEM 2009 mNarK. Positive feedback loop promoter, LasR regulated. Terminator. Kanamycin backbone for assembly. Chloramphenicol backbone for assembly. Amp&Kana backbone as back-up plan for assembly. Amp&Chlor backbone as back-up plan for assembly.
The missing important parts we need for ligation
RBS + coding region of LasR + terminator Red dye pathway (or any other colour) Native hypoxia promoter NarK.


 "
"




 Twitter
Twitter Facebook
Facebook UCL
UCL Flickr
Flickr YouTube
YouTube