Team:UIUC-Illinois/Modeling
From 2010.igem.org
(→Arsenic Accumulation) |
|||
| (17 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{Template:UIUC_Illinois_Bioware}} | ||
{{Template:UIUC_Illinois_Bioware_Background}} | {{Template:UIUC_Illinois_Bioware_Background}} | ||
| - | |||
| - | + | <br> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | == Cell Survivability == | ||
| + | '''This model demonstrates that the ''E. coli'' in our bioremediation system will have stable growth'''<br> | ||
| + | We have collaborated with the UIUC Software Tools team to analyze the ability of cells to survive when under the slight strain of overexpressing ArsB, ArsR, and LamB for the Arsenic Bioremediation system. Below is a summary of the work that Meagan Musselman and Anna Kropornica have compiled to demonstrate that indeed, ''E. coli'' cells will be well able to survive while over-expressing the aforementioned proteins.<br> | ||
| - | < | + | '''Modeling for UIUC Bioware''' |
| + | <br> | ||
| + | The Illinois Bioware team’s project was to develop bacteria able to facilitate the removal of heavy metals, a process known as bioremediation. The metal that was focused on was Arsenic. To test the program as well as to aid the Illinois Software team in their experimental procedure, we utilized the Constraint-based reconstruction analysis software1 through MATLAB to provide a prediction of growth for the Escherichia coli cells with the proteins implemented. In this modeling, we made the assumptions that the primary protein additions are the Arsenite/antimonite transporter (ArsB), DNA-binding transcriptional repressor (ArsR) and maltose outer membrane porin (LamB) proteins are the most influential components of the team’s Escherichia coli strain when in a medium containing the heavy metal, arsenic. The ArsB protein functions as a membrane transporting pump of arsenate, arsenite and antimonite outside of the cell while utilizing ATP4. The ArsR transcriptional repressor functions within a cell by regulating the expression of the arsRBC that is directly involved in creating resistance to arsenic4. Finally, the LamB protein functions as a maltoporin-like protein by transporting maltodextrins across the membrane2. <br> | ||
| + | The uptake rate of arsenic by ArsB was determined by Meng, et al. (2004). The dynamic flux balance analysis result (Figure 1) was determined with a initial concentration of 10-3 μmol and a initial biomass of 0.1 milligrams of Dry Weight Liter-1, a time of one hour and maximum number of steps at 20. In Figure 1, the plot shows changes in biomass concentration as a function of time on the left side, given the additional components. <br><br> | ||
| + | '''Figure 1'''<br> | ||
| + | [[Image:UIUC-IllinoisCellSurvivability.jpeg|500px]] | ||
| + | <br> | ||
| + | '''References''' | ||
| + | 1. Becker, Scott; et al.”Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox.” Department of Bioengineering, University of California San Diego.Nature Protocols Vol.2 No.3. March 2007.<br> | ||
| + | 2. Durfee T, et al. “The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse.” Bacteriol, 2008 Apr. <br> | ||
| + | 3. Meng, Y., et al. “As(III) and Sb(III) Uptake by GlpF and Efflux by ArsB in Escherichia coli.” The Journal of Biological Chemistry, 2004 Apr 30.<br> | ||
| + | 4. Ogura Y, et al. “Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli.” Ogura Y, et al. Proc Natl Acad Sci U S A, 2009 Oct 20. <br> | ||
| + | |||
| + | == Arsenic Accumulation == | ||
| + | '''This model demonstrates strong arsenic removal based on cell concentration and affinity for arsenic''' | ||
| + | <br><br> | ||
| + | '''Description'''<br> | ||
| + | Previous studies have found the arsenic accumulation by E. coli cells to be 1.47nmol/mg [dry weight] for E. coli producing phytochelatins and up to 2.3 nmol/mg [dry weight] for E. coli in which ArsR is expressed1. These numbers provided a reasonable range for the expected accumulation of our system. It was useful to note that the 2nmol/mg (dry weight) is approximately 50 μM arsenite which is approximately 6.5ppb (1ppb = 1μg/L). There was assumed to be a linear relationship between concentration of cells and arsenic accumulation. This is reasonable because the solution is assumed to be well mixed so that despite varying concentrations of cells, all cells are able to encounter equivalent amounts of arsenic. The assumption of the linear relationship makes it possible to model the system using a simple equation of the form y = m*x. The y-intercept is assumed to be zero because if no cells are present, no arsenic can be accumulated. The value m (or C in ArsCollection) represents the binding affinity and accumulation of arsenic due to the bacteria. This value can be altered to show what would be a reasonable concentration of cells to collect a given Arsenic concentration. In ArsCollection, m is graphed at 2, 4, 6, 8, and 10. A slope value of 6 most closely represents the accumulation seen in E. coli with over expression of ArsR under the T7 promoter1. Targeting the ArsR proteins to the cell surface allows for a significantly greater accumulation of arsenic. According to this model, the projected Arsenic collection for our system is well over 50ppb, particularly at cell concentrations over 7mg/mL. This is very encouraging and supports the success and effectiveness of our designed system. This model does not account for saturation that would occur once cells have accumulated the majority of the arsenic in the system. | ||
| + | <br><br> | ||
| + | Check for reasonability: Assuming 10^8 bacteria per mL3 and 20,000 proteins per cell2 and approximately 1.5 arsenic molecules bound to a given protein, the resulting amount of arsenic collected by the culture is: | ||
| + | <br> | ||
| + | (〖10〗^8 bacteria)/mL*20,000 proteins/cell*1.5 molecules/protein=3.0*(〖10〗^11 (arsenic molecules))/ml=0.5nM Arsenic≅0.13 ppb | ||
| + | <br> | ||
| + | Although 0.13 ppb is smaller than even the US standard of 10ppb, there are several possible areas in which the above values could be altered, and the result is still relatively similar to the expected value. In particular, an effective system would likely have the significantly more bacteria/mL. | ||
| + | <br><br> | ||
| + | '''Figure 1'''This image shows that the projected Arsenic collection for our system is well over 50ppb, particularly at cell concentrations over 7mg/mL.'''<br> | ||
| + | [[Image:UIUC-IllinoisAsCollection.jpg]]<br><br> | ||
| + | '''Program''' | ||
| + | The following is the function used in Matlab to produce the model and evaluate the data. Explanations are included in the comments.Here is a copy of the file. [[Media:UIUC-IllinoisArsCollection.m]]<br><br> | ||
| + | |||
| + | function y = ArsCollection<br> | ||
| + | %Assuming linear relationship between cell amount and amount of As<br> | ||
| + | %collected.<br> | ||
| + | x = 0:1/1000:10;<br> | ||
| + | for C = 2:2:10;<br> | ||
| + | %C represents cell affinity for As.<br> | ||
| + | %With arsenic naturally up-regulated in Wild Type cells this value is ~ 6<br> | ||
| + | %(Enhanced Arsenic Accumulation in Engineered Bacterial Cells <br> | ||
| + | %Expressing ArsR by Jan Kostal et. all.). <br> | ||
| + | %In our system, we would expect a value closer to 8 or 10 because<br> | ||
| + | %ArsR is being expressed on the membrane.<br> | ||
| + | y(:,C) = C.*x; xlabel('Dry Weight of Cells (mg/mL)'); ylabel('Arsenic Collection (ppb)');<br> | ||
| + | title('Arsenic Collection based on Cell Concentration and As Affinity');<br> | ||
| + | end<br> | ||
| + | plot(x,y);<br> | ||
| + | %Y axis in ppb collected.<br> | ||
| + | %X axis is mg of cells (dry weight) / mL | ||
| + | <br><br> | ||
| + | '''References'''<br> | ||
| + | |||
| + | 1. [http://aem.asm.org/cgi/content/abstract/70/8/4582 “Enhanced Arsenic Accumulation in Engineered Bacterial Cells Expressing ArsR”]. Jan Kostal,1 Rosanna Yang,1 Cindy H. Wu,1,2 Ashok Mulchandani,1 and Wilfred Chen1*<br> | ||
| + | 2. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC215663/pdf/jbacter00231-0009.pdf “High-Sensitivity Detection of Newly Induced LamB Protein on the Escherichia coli Cell Surface”]. G. H. VOS-SCHEPERKEUTER,1 M; HOFNUNG,2 AND B. WITHOLTl*<br> | ||
| + | 3. [http://jb.asm.org/cgi/content/full/189/23/8746 “Escherichia coli Physiology in Luria-Bertani Broth”] | ||
| + | Guennadi Sezonov,* Danièle Joseleau-Petit, and Richard D'Ari | ||
| + | Institut Jacques Monod.<br> | ||
Latest revision as of 03:42, 28 October 2010
Cell Survivability
This model demonstrates that the E. coli in our bioremediation system will have stable growth
We have collaborated with the UIUC Software Tools team to analyze the ability of cells to survive when under the slight strain of overexpressing ArsB, ArsR, and LamB for the Arsenic Bioremediation system. Below is a summary of the work that Meagan Musselman and Anna Kropornica have compiled to demonstrate that indeed, E. coli cells will be well able to survive while over-expressing the aforementioned proteins.
Modeling for UIUC Bioware
The Illinois Bioware team’s project was to develop bacteria able to facilitate the removal of heavy metals, a process known as bioremediation. The metal that was focused on was Arsenic. To test the program as well as to aid the Illinois Software team in their experimental procedure, we utilized the Constraint-based reconstruction analysis software1 through MATLAB to provide a prediction of growth for the Escherichia coli cells with the proteins implemented. In this modeling, we made the assumptions that the primary protein additions are the Arsenite/antimonite transporter (ArsB), DNA-binding transcriptional repressor (ArsR) and maltose outer membrane porin (LamB) proteins are the most influential components of the team’s Escherichia coli strain when in a medium containing the heavy metal, arsenic. The ArsB protein functions as a membrane transporting pump of arsenate, arsenite and antimonite outside of the cell while utilizing ATP4. The ArsR transcriptional repressor functions within a cell by regulating the expression of the arsRBC that is directly involved in creating resistance to arsenic4. Finally, the LamB protein functions as a maltoporin-like protein by transporting maltodextrins across the membrane2.
The uptake rate of arsenic by ArsB was determined by Meng, et al. (2004). The dynamic flux balance analysis result (Figure 1) was determined with a initial concentration of 10-3 μmol and a initial biomass of 0.1 milligrams of Dry Weight Liter-1, a time of one hour and maximum number of steps at 20. In Figure 1, the plot shows changes in biomass concentration as a function of time on the left side, given the additional components.
Figure 1
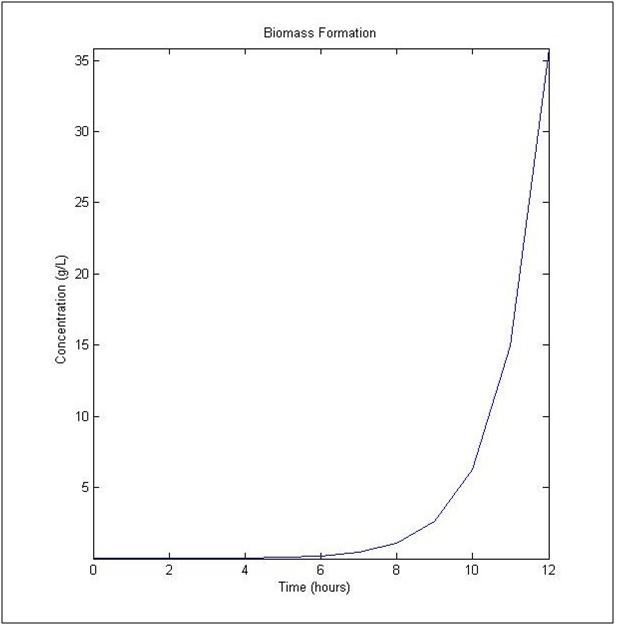
References
1. Becker, Scott; et al.”Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox.” Department of Bioengineering, University of California San Diego.Nature Protocols Vol.2 No.3. March 2007.
2. Durfee T, et al. “The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse.” Bacteriol, 2008 Apr.
3. Meng, Y., et al. “As(III) and Sb(III) Uptake by GlpF and Efflux by ArsB in Escherichia coli.” The Journal of Biological Chemistry, 2004 Apr 30.
4. Ogura Y, et al. “Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli.” Ogura Y, et al. Proc Natl Acad Sci U S A, 2009 Oct 20.
Arsenic Accumulation
This model demonstrates strong arsenic removal based on cell concentration and affinity for arsenic
Description
Previous studies have found the arsenic accumulation by E. coli cells to be 1.47nmol/mg [dry weight] for E. coli producing phytochelatins and up to 2.3 nmol/mg [dry weight] for E. coli in which ArsR is expressed1. These numbers provided a reasonable range for the expected accumulation of our system. It was useful to note that the 2nmol/mg (dry weight) is approximately 50 μM arsenite which is approximately 6.5ppb (1ppb = 1μg/L). There was assumed to be a linear relationship between concentration of cells and arsenic accumulation. This is reasonable because the solution is assumed to be well mixed so that despite varying concentrations of cells, all cells are able to encounter equivalent amounts of arsenic. The assumption of the linear relationship makes it possible to model the system using a simple equation of the form y = m*x. The y-intercept is assumed to be zero because if no cells are present, no arsenic can be accumulated. The value m (or C in ArsCollection) represents the binding affinity and accumulation of arsenic due to the bacteria. This value can be altered to show what would be a reasonable concentration of cells to collect a given Arsenic concentration. In ArsCollection, m is graphed at 2, 4, 6, 8, and 10. A slope value of 6 most closely represents the accumulation seen in E. coli with over expression of ArsR under the T7 promoter1. Targeting the ArsR proteins to the cell surface allows for a significantly greater accumulation of arsenic. According to this model, the projected Arsenic collection for our system is well over 50ppb, particularly at cell concentrations over 7mg/mL. This is very encouraging and supports the success and effectiveness of our designed system. This model does not account for saturation that would occur once cells have accumulated the majority of the arsenic in the system.
Check for reasonability: Assuming 10^8 bacteria per mL3 and 20,000 proteins per cell2 and approximately 1.5 arsenic molecules bound to a given protein, the resulting amount of arsenic collected by the culture is:
(〖10〗^8 bacteria)/mL*20,000 proteins/cell*1.5 molecules/protein=3.0*(〖10〗^11 (arsenic molecules))/ml=0.5nM Arsenic≅0.13 ppb
Although 0.13 ppb is smaller than even the US standard of 10ppb, there are several possible areas in which the above values could be altered, and the result is still relatively similar to the expected value. In particular, an effective system would likely have the significantly more bacteria/mL.
Figure 1This image shows that the projected Arsenic collection for our system is well over 50ppb, particularly at cell concentrations over 7mg/mL.
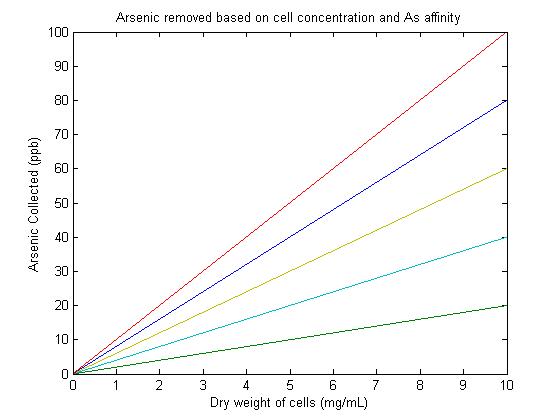
Program
The following is the function used in Matlab to produce the model and evaluate the data. Explanations are included in the comments.Here is a copy of the file. Media:UIUC-IllinoisArsCollection.m
function y = ArsCollection
%Assuming linear relationship between cell amount and amount of As
%collected.
x = 0:1/1000:10;
for C = 2:2:10;
%C represents cell affinity for As.
%With arsenic naturally up-regulated in Wild Type cells this value is ~ 6
%(Enhanced Arsenic Accumulation in Engineered Bacterial Cells
%Expressing ArsR by Jan Kostal et. all.).
%In our system, we would expect a value closer to 8 or 10 because
%ArsR is being expressed on the membrane.
y(:,C) = C.*x; xlabel('Dry Weight of Cells (mg/mL)'); ylabel('Arsenic Collection (ppb)');
title('Arsenic Collection based on Cell Concentration and As Affinity');
end
plot(x,y);
%Y axis in ppb collected.
%X axis is mg of cells (dry weight) / mL
References
1. [http://aem.asm.org/cgi/content/abstract/70/8/4582 “Enhanced Arsenic Accumulation in Engineered Bacterial Cells Expressing ArsR”]. Jan Kostal,1 Rosanna Yang,1 Cindy H. Wu,1,2 Ashok Mulchandani,1 and Wilfred Chen1*
2. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC215663/pdf/jbacter00231-0009.pdf “High-Sensitivity Detection of Newly Induced LamB Protein on the Escherichia coli Cell Surface”]. G. H. VOS-SCHEPERKEUTER,1 M; HOFNUNG,2 AND B. WITHOLTl*
3. [http://jb.asm.org/cgi/content/full/189/23/8746 “Escherichia coli Physiology in Luria-Bertani Broth”]
Guennadi Sezonov,* Danièle Joseleau-Petit, and Richard D'Ari
Institut Jacques Monod.
 "
"
