|
|
| (86 intermediate revisions not shown) |
| Line 2: |
Line 2: |
| | {{ETHZ_Basel10_Modeling}} | | {{ETHZ_Basel10_Modeling}} |
| | | | |
| - | = Molecular Modeling Overview = | + | = Mathematical Modeling Overview = |
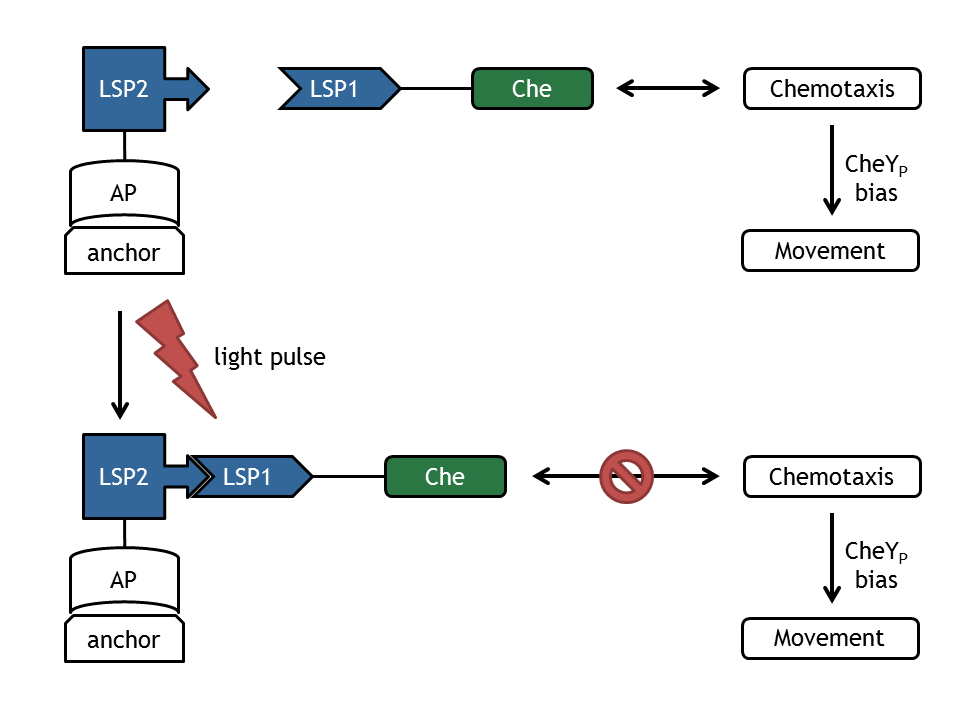
| | + | [[Image:ETHZ_Basel_molecular_comb.png|thumb|400px|'''Figure 1: schematical overview of the modeled processes in E. lemming.''' LSP refers to light switch protein, AP to anchor protein, and Che to the attacked protein of the chemotaxis pathway.]] |
| | | | |
| | + | A complex mathematical model of E. lemming from both literature inspired and self developed submodels was created that covers the processes displayed in Figure 1. |
| | | | |
| - | ETHZ’s project, E-Lemming, aims to modify the chemotaxis property of E.coli such that, instead of response to a chemical attractant/repellent, the bacterium responds to a light stimulus (phototaxis). Furthermore, this light sensitivity is used to control E.coli’s movement by deciding, at any given time, which type of motion will our ‘Lemming – in – disguise’ adopt (tumbling or straight run). This leads to a controllable E.coli, which can follow any pre – defined spatial path/maze/game, as a result of the combination of tumbling and going straight. The bacterium colony is imaged and, by image processing, the position of a single tracked cell is inferred. By activating a light switch, the user decides whether the bacterium should continue running or should change direction.
| + | In a first step, existing models for the individual processes of E. lemming have been identified by literature research, implemented, corrected and adapted to our needs. Where we could not rely on established models, we started modeling on our own and calibrated the model with regard to available literature knowledge. |
| - | <br><br>
| + | |
| - | In the theory world, the steps we are following in mindlessly driving E.coli to our pre - defined target are the following:
| + | |
| | | | |
| - | • deterministic (ODE) & stochastic models of the chemotaxis pathway (documented from the literature)<br>
| + | * [[Team:ETHZ_Basel/Modeling/Light_Switch|'''Light Switch''']]: both implementation approaches have been modeled: |
| - | • model of the movement of E.coli (built on the information of the pathway derived from the molecular models) <br>
| + | ** [[Team:ETHZ_Basel/Modeling/Light_Switch#Modeling_of_the_light_switch:_PhyB.2FPIF3|'''PhyB/PIF3''']]: a deterministic molecular model based on the light-sensitive dimerizing Arabidopsis proteins PhyB and PIF3. |
| - | • control algorithms ( built on the user’s desire to play around with E.coli)<br>
| + | ** [[Team:ETHZ_Basel/Modeling/Light_Switch#Modeling_of_the_PhyB.2FPIF3_light_switch#Archeal_light_receptor|'''Archeal Light Receptor''']]: a deterministic molecular model based on the archeal light receptor. |
| - | • image tracking & image processing algorithms<br>
| + | * [[Team:ETHZ_Basel/Modeling/Chemotaxis|'''Chemotaxis Pathway''']]: two deterministic molecular models of the chemotaxis pathway. |
| - | • java applications/movies of the E.Lemming (the fun part)<br>
| + | * [[Team:ETHZ_Basel/Modeling/Movement|'''Bacterial Movement''']]: a self developed stochastic model of ''E. coli'' movement on basis of the CheYp bias. |
| | | | |
| - | == The network topology==
| + | In a second part, we combined the submodels stepwise to more comprehensive models that we could use to address different important questions to: |
| - | | + | * [[Team:ETHZ_Basel/Modeling/Combined#PhyB.2FPIF3_light_switch_-_Chemotaxis |'''PhyB/PIF3 light switch - Chemotaxis''']]: this model was used to reduce [[Team:ETHZ_Basel/Biology|wet laboratory experiments]] by identification molecular targets by [[Team:ETHZ_Basel/Modeling/Experimental_Design|experimental design]]. |
| - | Many bacteria posses a signal transduction network that makes it possible to sense the changes in concentration of a certain chemical attractant/repellent in the extracellular environment and direct the movement towards the attractant and away from the repellent. This property is what is known as ‘''chemotaxis''’. There are basically two types of movement that the bacterium can employ: ''''tumbling'''', which means change of direction and occurs when the bacterium doesn’t sense an increase of attractant concentration in the extracellular environment anymore and ''''running straight'''', which occurs when the attractant concentration is increasing. The running straight stands for “correct direction, ok, keep going” (towards the attractant/away from repellent) while the tumbling stands for “wrong direction, give it another shot”. In the case of E.coli, these two types of movements correspond to different rotation directions of its 4 flagellar motors (clockwise: tumbling; counter-clockwise: runs).<br>
| + | * [[Team:ETHZ_Basel/Modeling/Combined#Archeal_light_receptor_-_Chemotaxis |'''Archeal light receptor - Chemotaxis''']]: this model was combined identically to the one above. |
| - | The direction of rotation of the motors can be determined by the ratio of concentrations of certain proteins inside the cell. That is, there are some quantitative indicators from which one can infer whether the bacterium will run or it will go straight, as a response to changes in input concentration. These indicators (concentration levels for certain proteins) are the result of a well – studied signal transduction network, consisting of membrane receptor proteins and intracellular proteins (Che). <br>
| + | * [[Team:ETHZ_Basel/Modeling/Combined#Chemotaxis_-_Movement |'''Chemotaxis - Movement''']]: complete model of E. lemming as a simulative test bench for the [[Team:ETHZ_Basel/InformationProcessing/Controller|controller]] design and as a brick of the comprehensive simulation of [[Team:ETHZ_Basel/InformationProcessing|information processing]]. |
| - | Our models were constructed based on the following network topology: <br><br>
| + | |
| - | <no worries, picture to be inserted><br><br><br>
| + | |
| - | | + | |
| - | CheW and CheA are localized right next to the membrane, inside the cell. They form a complex together with the receptor clusters on the membrane (which sense the changes in input concentration in the extracellular environment). This complex can be successively methylated (on different methylation states), a constant process done by CheR. In the literature, there are usually 5 methylation sites considered (m = 0,1,2,3,4; m=0 stands for no methylation and m=4 stands for the highest methylation level). <br>
| + | |
| - | The key process that influences the outcome of the network (tumbling/running) is the autophosphorylation of the protein CheA, which is favoured either by the increase in the methylation state of the membrane complex, done by CheR, or by decreasing attractant concentration.<br>
| + | |
| - | With decreased attractant (increased repellent) concentration, CheA is autophosphorylated. The phosphoryl groups are being used to produce either CheYp or CheBp. CheYp diffuses through the cytoplasm to the motors, generating clockwise spins, therefore tumbling. CheBp is responsible for de-methylation of the receptors, which reduces the chances of CheA being autophosphorylated, therefore the system is returning to its previous state.<br>
| + | |
| - | With increased attractant (decreased repellent) concentration, the rate of CheA autophosphorylation is reduced, meaning that there are less phosphate groups available for CheYp and CheBp. Therefore the level of CheYp decreases and the level of CheY increases, which leads to straight runs. Then, the level of CheBp decreases, increasing the chances of CheA being autophosphorylated (since the receptors are methylated by CheR), therefore returning the system again to its previous state.<br><br>
| + | |
| - | | + | |
| - | A perfect model of the chemotaxis pathway should reconstruct a system that exhibits the following characteristics:<br>
| + | |
| - | • '''adaptation''': the ability to respond to changes in the external environment and return the intracellular protein phosphorylation levels to their pre-stimulus levels.<br>
| + | |
| - | • '''sensitivity''': even small changes in the concentration of the attractant/repellent trigger chemotactic responses from bacteria.<br>
| + | |
| - | • '''gain''': the ability to amplify the received signal from the extracellular environment, so as to modulate the intracellular signaling cascade.<br>
| + | |
| - | • '''robustness''': perturbations of the network’s parameters should not influence the behavior of the system.<br><br>
| + | |
| - | Even though the chemotaxis system has been long and in depth studied and it is well documented in the literature (from a modeling point of view), it is to be noted that none of the existing models, accounting for experimental findings, can satisfy all of the above requirments. So, we decided to implement several models to answer our questions.
| + | |
| - | | + | |
| - | == Model coupling ==
| + | |
| - | | + | |
| - | [[Image:Modeling_overview.png|center|600px|Coupled models for the simulation of the whole process and their interfaces.]] | + | |
| - | | + | |
| - | Besides the ''in vivo'' setup allowing the steering of a single ''Escherichia coli'' cell, the whole setup will be modeled. This allows us the testing of the cell detection & tracking as well as the design of the control algorithm in order to start verifying the different biological constructs and to record the successful control of bacterial movement.
| + | |
A complex mathematical model of E. lemming from both literature inspired and self developed submodels was created that covers the processes displayed in Figure 1.
In a first step, existing models for the individual processes of E. lemming have been identified by literature research, implemented, corrected and adapted to our needs. Where we could not rely on established models, we started modeling on our own and calibrated the model with regard to available literature knowledge.
In a second part, we combined the submodels stepwise to more comprehensive models that we could use to address different important questions to:
 "
"