Team:ETHZ Basel/InformationProcessing/Visualization
From 2010.igem.org
(→Visualization) |
(→User Experience) |
||
| (25 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
{{ETHZ_Basel10_InformationProcessing}} | {{ETHZ_Basel10_InformationProcessing}} | ||
| - | = Visualization = | + | = Visualization and Man-Machine Interface = |
| - | == | + | == Background == |
| - | + | <html><div class="thumb tright"><div class="thumbinner" style="width:402px;"> | |
| + | <iframe title="YouTube video player" class="youtube-player" type="text/html" width="400" height="325" src="http://www.youtube.com/embed/hwtOBgQCAAA?rel=0&hd=1" frameborder="0"></iframe> | ||
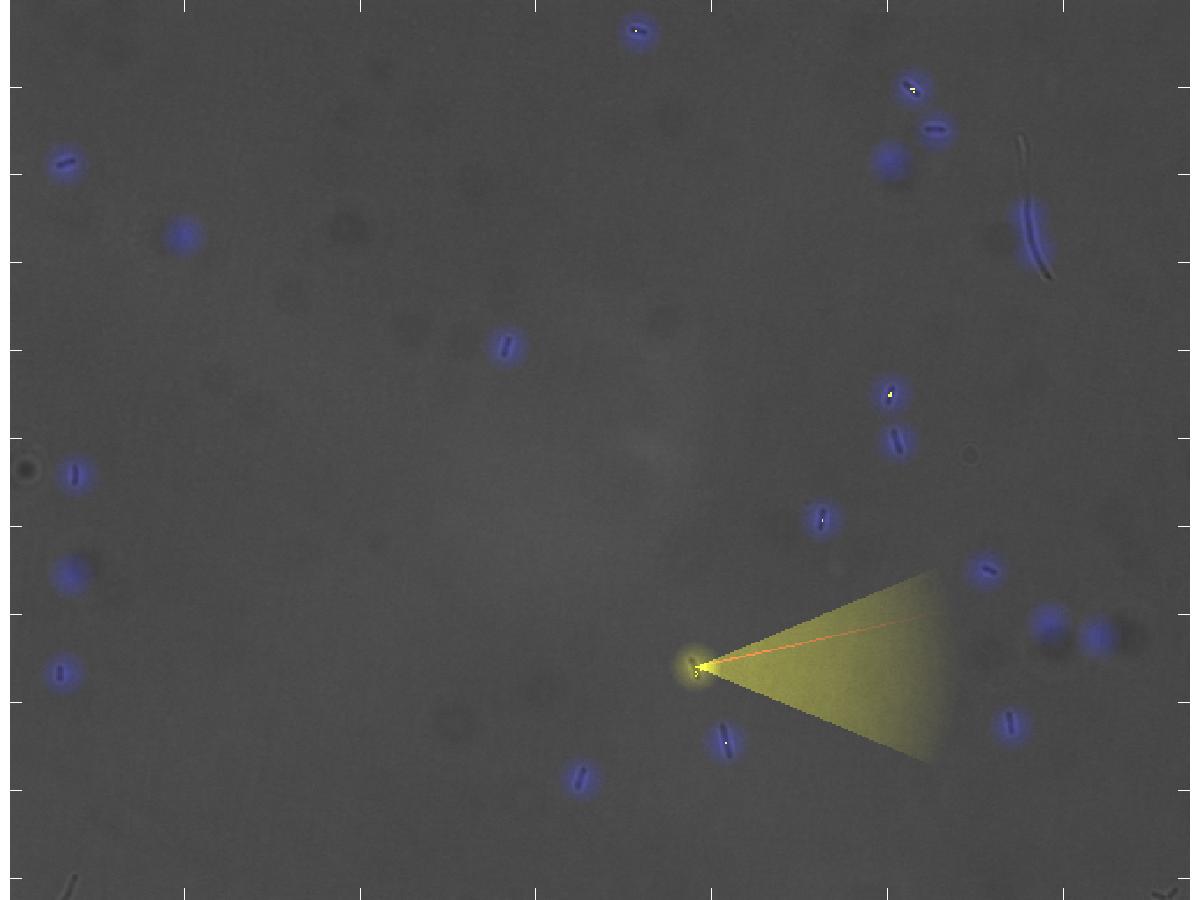
| + | <div class="thumbcaption"><div class="magnify"><a href="http://www.youtube.com/watch?v=hwtOBgQCAAA&hd=1" class="external" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div><b>The real-time representation of the current microscope image</b>. <br><i>Blue dots</i>: the detected <i>E. coli</i> cells. <i>Yellow dot</i>: the currently selected E. lemming. <i>Yellow cone</i>: the current swimming direction of E. lemming. <i>Red thin line</i>: the reference direction. <i>Yellow dotted line</i>: the current path of the E. lemming. <br>Note that the movement process is not under the influence of the controller.</br> </div></div></div></html> | ||
| + | To control E. lemming, the scientist has to be aware of a huge amount of information, which has to be visualized in real-time. Only with this information, the user is able to choose an E. lemming to control, decide on a reference direction, and interfere in the experiments. This information includes | ||
| + | * the current microscopy image; | ||
| + | * all detected, not detected and falsely detected cells; | ||
| + | * the selected cell itself; | ||
| + | * its swimming direction; | ||
| + | * the reference direction; | ||
| + | * its path; | ||
| + | * and movements of the stage. | ||
| + | Since this information has to be absorbed by the experimentalist at a rate higher than three frames per second, a textual representation is not adequate. We thus decided to represent the information in an intuitive way by incorporating it into the microscope images. | ||
| - | + | To enable the scientist to interfere with the experiment without having to interrupt, and thus possibly losing the currently controlled cell, we established two input devices: the user can either control the experiment and the E. lemming using the keyboard or the joystick. With both devices he/she is able to: | |
| + | * select a cell to control; | ||
| + | * activate or deactivate the controller; | ||
| + | * set the reference direction; | ||
| + | * increase or decrease the threshold for cell detection; | ||
| + | * or, alternatively, manually induce red or far-red light pulses of various length (instead of using the controllers provided in the toolbox). | ||
| + | If using a force-feedback joystick, the current swimming direction is furthermore given as a small force on the direction of the joystick, thus intuitively increasing the amount of information available for the user. | ||
| - | + | Both, the visual representation of the current state of the experiment as well as the user input had to be designed to require low computational efforts not to unnecessarily slow down the imaging. | |
| - | == | + | == User Experience == |
| - | + | [[Image:userExperience.jpg|thumb|400px|'''Example Image of the Visualization''']]The images of the microscope are visualized live at a personal computer which is connected to the microscope over a local network or internet, thus it is possible to control the E. lemming from another office or city. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | === Visual Enhancements === | |
| - | The | + | All detected cells in the microscope image are highlighted by letting them glow blue (see example picture). The user can select a cell to control with the joystick or the keyboard, which is then glowing yellow instead of blue. The current swimming direction is indicated by a transparent yellow light cone, imitating the floodlight of cars or a flashlight, and the reference direction is depicted as a thin red line. The path the cell has passed during the experiment is marked by small yellow dots, similar to footprints in snow. Finally, the automatic movement of the stage of the microscope, when following a cell which swims out of the current field of view, is indicated by arrows in the direction of movement, which are popping up for a moment. |
Latest revision as of 01:13, 28 October 2010
Visualization and Man-Machine Interface
Background
Blue dots: the detected E. coli cells. Yellow dot: the currently selected E. lemming. Yellow cone: the current swimming direction of E. lemming. Red thin line: the reference direction. Yellow dotted line: the current path of the E. lemming.
Note that the movement process is not under the influence of the controller.
- the current microscopy image;
- all detected, not detected and falsely detected cells;
- the selected cell itself;
- its swimming direction;
- the reference direction;
- its path;
- and movements of the stage.
Since this information has to be absorbed by the experimentalist at a rate higher than three frames per second, a textual representation is not adequate. We thus decided to represent the information in an intuitive way by incorporating it into the microscope images.
To enable the scientist to interfere with the experiment without having to interrupt, and thus possibly losing the currently controlled cell, we established two input devices: the user can either control the experiment and the E. lemming using the keyboard or the joystick. With both devices he/she is able to:
- select a cell to control;
- activate or deactivate the controller;
- set the reference direction;
- increase or decrease the threshold for cell detection;
- or, alternatively, manually induce red or far-red light pulses of various length (instead of using the controllers provided in the toolbox).
If using a force-feedback joystick, the current swimming direction is furthermore given as a small force on the direction of the joystick, thus intuitively increasing the amount of information available for the user.
Both, the visual representation of the current state of the experiment as well as the user input had to be designed to require low computational efforts not to unnecessarily slow down the imaging.
User Experience
The images of the microscope are visualized live at a personal computer which is connected to the microscope over a local network or internet, thus it is possible to control the E. lemming from another office or city.Visual Enhancements
All detected cells in the microscope image are highlighted by letting them glow blue (see example picture). The user can select a cell to control with the joystick or the keyboard, which is then glowing yellow instead of blue. The current swimming direction is indicated by a transparent yellow light cone, imitating the floodlight of cars or a flashlight, and the reference direction is depicted as a thin red line. The path the cell has passed during the experiment is marked by small yellow dots, similar to footprints in snow. Finally, the automatic movement of the stage of the microscope, when following a cell which swims out of the current field of view, is indicated by arrows in the direction of movement, which are popping up for a moment.
 "
"