Team:HokkaidoU Japan/Notebook/September15
From 2010.igem.org
(→Result of yesterdays transformation) |
|||
| (17 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{Template:HokkaidoU_Japan}} | + | {{Template:HokkaidoU_Japan}}<div class="linkbar"><div class="prev">[[Team:HokkaidoU_Japan/Notebook/September14|September 14]]</div>[[Team:HokkaidoU_Japan/Notebook|Notebook]]<div class="next">[[Team:HokkaidoU_Japan/Notebook/September17|September 17]]</div></div> |
| + | |||
*Observed results of yesterdays transformation | *Observed results of yesterdays transformation | ||
**Transformation using heat shock went well | **Transformation using heat shock went well | ||
| Line 8: | Line 9: | ||
| - | == Result of yesterdays transformation == | + | == Result of yesterdays [[Team:HokkaidoU_Japan/Protocols|transformation]] == |
Transformation using heat shock went well and produced about 100 colonies. We chose 10 and did colony PCR to check if insert was correct. The insert was Arabinose promoter[http://partsregistry.org/Part:BBa_I0500 (BBa_I0500)] and GFP reporter[http://partsregistry.org/Part:BBa_E0840 (BBa_E0840)]. Vector used was [http://partsregistry.org/wiki/index.php?title=Part:pSB1C3 pSB1C3]. | Transformation using heat shock went well and produced about 100 colonies. We chose 10 and did colony PCR to check if insert was correct. The insert was Arabinose promoter[http://partsregistry.org/Part:BBa_I0500 (BBa_I0500)] and GFP reporter[http://partsregistry.org/Part:BBa_E0840 (BBa_E0840)]. Vector used was [http://partsregistry.org/wiki/index.php?title=Part:pSB1C3 pSB1C3]. | ||
| - | [[Image:HokkaidoU_Pictures_Colony_PCR_of_2010_09_15.png| | + | <br> |
| + | |||
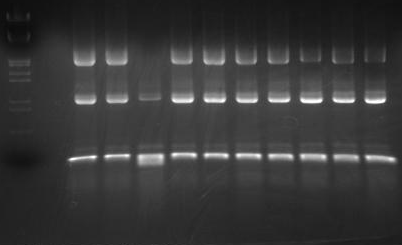
| + | [[Image:HokkaidoU_Pictures_Colony_PCR_of_2010_09_15.png|200px|right|thumb|Electrophoresis of colony PCR]] | ||
| + | |||
| + | Combined length of insert with prefix, suffix and scar is estimated to be 2143bp. From electrophoresis picture under UV light you can see all colonies except colony-3 have bands at approximate 2000bp position. | ||
| + | |||
| + | | ||
| + | |||
| + | We used DNA marker [https://2010.igem.org/Image:HokkaidoU_Pictures_DNA_Marker.png Lambda/''Hin''dIII, EcoR I] followings are empty lane and then colonies 1 through 10 accordingly. We used 1.5% agarose gel for electrophoresis. | ||
| + | |||
| + | | ||
| + | |||
| + | Unfortunately electroporation transformation failed to produce colonies. This was due to fact that competent cells weren't prepared for electroporation. We used cells prepared for heat shock transformation. | ||
| + | |||
| + | == Incubation in L(+)Arabinose medium == | ||
| + | |||
| + | We added other half of E.Coli cell used in colony PCR to L(+)Arabinose medium to check phenotype. Also did L(+)Arabinose negative control for all the colonies, GFP (-) L(+) Arabinose (+), GFP (-) L(+) Arabinose (-) and E.Coli (-) controls. | ||
| + | |||
| + | | ||
| + | |||
| + | Medium was inoculated with Chloramphenicol. GFP (-) E.coli had pUC119 plasmid so medium was Ampicilin inoculated. | ||
| - | + | == Transformation of Arabinose promoter == | |
| - | + | Part [http://partsregistry.org/Part:BBa_I0500 (BBa_I0500)] is necessary for our T3SS project so we decided to make glycerol because one of other promoters recently dried up and became unusable. | |
Latest revision as of 08:25, 27 October 2010
- Observed results of yesterdays transformation
- Transformation using heat shock went well
- Electroporation transformation failed produce colonies
- Did Colony PCR of yesterdays transformed colonies
- Introduced colonies to L(+)Arabinose medium to check if it would show desired function
- Check for results tomorrow
Result of yesterdays transformation
Transformation using heat shock went well and produced about 100 colonies. We chose 10 and did colony PCR to check if insert was correct. The insert was Arabinose promoter[http://partsregistry.org/Part:BBa_I0500 (BBa_I0500)] and GFP reporter[http://partsregistry.org/Part:BBa_E0840 (BBa_E0840)]. Vector used was [http://partsregistry.org/wiki/index.php?title=Part:pSB1C3 pSB1C3].
Combined length of insert with prefix, suffix and scar is estimated to be 2143bp. From electrophoresis picture under UV light you can see all colonies except colony-3 have bands at approximate 2000bp position.
We used DNA marker Lambda/HindIII, EcoR I followings are empty lane and then colonies 1 through 10 accordingly. We used 1.5% agarose gel for electrophoresis.
Unfortunately electroporation transformation failed to produce colonies. This was due to fact that competent cells weren't prepared for electroporation. We used cells prepared for heat shock transformation.
Incubation in L(+)Arabinose medium
We added other half of E.Coli cell used in colony PCR to L(+)Arabinose medium to check phenotype. Also did L(+)Arabinose negative control for all the colonies, GFP (-) L(+) Arabinose (+), GFP (-) L(+) Arabinose (-) and E.Coli (-) controls.
Medium was inoculated with Chloramphenicol. GFP (-) E.coli had pUC119 plasmid so medium was Ampicilin inoculated.
 "
"