Team:Michigan/Alex's Notebook
From 2010.igem.org
m (→10/3/10) |
|||
| (6 intermediate revisions not shown) | |||
| Line 546: | Line 546: | ||
*2 mL LB in 15mL tube | *2 mL LB in 15mL tube | ||
*37°C, 200 rpm shaking - ERB 1230 | *37°C, 200 rpm shaking - ERB 1230 | ||
| + | Uploaded NanR cloning [[media:NanR_Cloning.pdf|protocol]] | ||
--[[User:Infekt|Infekt]] 05:35, 13 September 2010 (UTC) | --[[User:Infekt|Infekt]] 05:35, 13 September 2010 (UTC) | ||
| + | |||
| + | == 9/29/10 == | ||
| + | ''Alex and John'' | ||
| + | |||
| + | Dissolved M9 salts to 50X | ||
| + | *All of component A in 1000 mL DIwater | ||
| + | *All of component B in 1000 mL DIwater | ||
| + | Autoclaved both: each split into two 1000mL bottles filled w/ 500mL each (four bottles total) | ||
| + | *30 min sterilize time | ||
| + | Began biofilm assay following [[Media:Static+biofilm+quantification.pdf|Alex's biofilm assay protocol]] | ||
| + | |||
| + | Made broth cultures of ''P. putida'' LD1, ''P. fluorescens'' LD2, and LD1+LD2 coculture from plates, and ''P. putida'' KT2440 from Lin Lab frozen stock | ||
| + | *KT2440 inoculated into 2 mL LB in 15mL tube | ||
| + | *others inoculated into 2 mL LB + 100μg/mL amp in 15mL tube | ||
| + | *all incubated in 30°C, 200rpm shaking -- ERB 1230, 8:45pm | ||
| + | Streaked KT2440 on LB plate from frozen | ||
| + | *incubated 37°C -- ERB 1239 | ||
| + | Added KT2440 to strain database | ||
| + | Mixed M9 A and B solutions and diluted 1/50 in 200 mL sterile DIwater -- in 4 50mL tubes | ||
| + | *each tube 1 mL M9A, 1 mL M9B and 48 mL water | ||
| + | ---- | ||
| + | ''Alex'' | ||
| + | |||
| + | Uploaded [[Media:Static+biofilm+quantification.pdf|Alex's biofilm assay protocol]] | ||
| + | |||
| + | Added borax to two of the M9 tubes (100 mL) to buffer at ~pH 9.2 | ||
| + | - (.01 mol/L)*(.05 L)*(381.37 g/mol) = 190.7 mg borax/50 mL water | ||
| + | *Tested pH with pH indicator strips | ||
| + | **~9 | ||
| + | *Vacuum-filtered 100 mL w/ steriflips | ||
| + | |||
| + | == 9/30/10 == | ||
| + | ''Alex'' | ||
| + | |||
| + | Aliquot M9 and buffered M9 into five 15mL tubes each | ||
| + | Need 15 mL stocks of each of the following media -- each 15 mL neutral and 15 mL buffered at pH 9: | ||
| + | *M9 | ||
| + | *M9 + 0.4% glucose | ||
| + | *M9 + Acros NAs (250mg/L) | ||
| + | *M9 + Sigma Aldrich NAs (250mg/L) | ||
| + | *M9 + both Acros & Sigma NAs (each 250mg/L) | ||
| + | *LB | ||
| + | Added 150 μL 40% glucose to appropriate tubes (.4% final concentration) | ||
| + | Aliquot 15 mL LB into each of two 15mL tubes | ||
| + | Added NAs to appropriate tubes | ||
| + | - Acros: (.25 mg/mL)*(15mL)/(.91 mg/μL) = '''4.12 μL''' | ||
| + | - Sigma: (.25 mg/mL)*(15mL)/(.92 mg/μL) = '''4.08 μL''' | ||
| + | *added 4.1 μL of each NA to each respective tube | ||
| + | Added borax to one tube of LB to buffer at pH9 | ||
| + | - (190.7 mg)*(15μL/50μL) = '''57.2 mg''' per 15mL LB | ||
| + | *tested pH with indicator strip | ||
| + | **~9 | ||
| + | Tested pH of neutral M9 with indicator strip | ||
| + | *~7 | ||
| + | Moved KT2440 LB plate from 37°C to 4°C -- ERB 1239 | ||
| + | |||
| + | Continued [[Media:Static+biofilm+quantification.pdf|Alex's biofilm assay protocol]] | ||
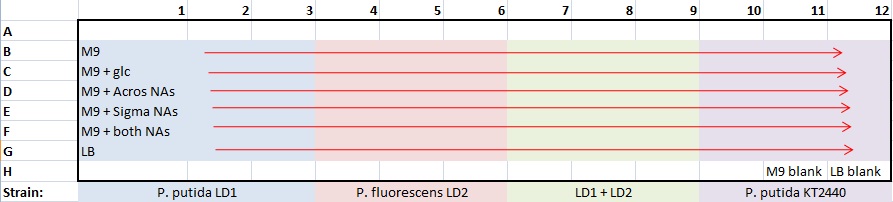
| + | *made one 96-well plate for all strains and media at pH7 only | ||
| + | *plate template : | ||
| + | [[image:9-30 plate template.jpg]] | ||
| + | **100 μL/well: 99 μL medium + 1 μL culture | ||
| + | **covered plate with gas-permeable membrane | ||
| + | *Started 24-hr kinetic reading in Lin Lab microplate reader | ||
| + | **30°C | ||
| + | **OD600 absorbance | ||
| + | **read every 30 min | ||
| + | Made broth cultures for LD1, LD2 and KT2440 from yesterday's overnight broths | ||
| + | *2 mL each in a 15mL tube | ||
| + | *30°C, 200 rpm shaking - '''6:20pm''' | ||
| + | Discarded yesterday's overnight broths | ||
| + | |||
| + | --[[User:Infekt|Infekt]] 00:57, 1 October 2010 (UTC) | ||
| + | |||
| + | == 10/1/10 == | ||
| + | ''Alex'' | ||
| + | |||
| + | Moved broth cultures 37°C to 4°C -- 1:30pm (19-hr culture) | ||
| + | Prepared biofilm assay plate for all pH9 tests - following [[Media:Static+biofilm+quantification.pdf|Alex's biofilm assay protocol]] | ||
| + | *plate template: | ||
| + | [[image:9-30 plate template.jpg]] | ||
| + | **100 μL/well: 99 μL medium + 1 μL culture | ||
| + | *stored plate in Lin Lab 4°C -- 2:20pm | ||
| + | ---- | ||
| + | Saved data for curves from ph7 plate | ||
| + | Read pH7 plate ODs | ||
| + | Performed biofilm assay on ph7 plate | ||
| + | Started 24-hr kinetic reading for pH9 plate -- ~midnight | ||
| + | |||
| + | Discarded pH7 plate | ||
| + | |||
| + | Processed pH7 plate data | ||
| + | *uploaded results [[Media:Ph7_biofilm_assay_results.xls|here]] | ||
| + | |||
| + | --[[User:Infekt|Infekt]] 06:55, 2 October 2010 (UTC) | ||
| + | |||
| + | Uploaded pH7 kinetic data [[Media:Data_10-01-10-pH7_kinetic.txt|here]] | ||
| + | |||
| + | --[[User:Infekt|Infekt]] 04:06, 4 October 2010 (UTC) | ||
| + | |||
| + | == 10/3/10 == | ||
| + | ''Alex'' | ||
| + | |||
| + | Ran CV biofilm assay on pH9 plate, following [[Media:Static+biofilm+quantification.pdf|my protocol]] | ||
| + | *discarded plate | ||
| + | *uploaded [[Media:Ph9_biofilm_assay_results.xls|results]] | ||
| + | *uploaded [[Media:Data_10-01-10-pH9_kinetic.txt|pH9 kinetic raw data]] | ||
| + | |||
| + | --[[User:Infekt|Infekt]] 04:35, 4 October 2010 (UTC) | ||
Latest revision as of 04:36, 4 October 2010
Alex's Wiki
7/7/2010
Jennifer and Alex - in Lin Lab
Made CaCl2 stock solution: 0.1M - 50mL
- molar mass = 111 g/mol
- (0.1 mol/L)(.05 L)(111 g/mol) = 555mg
- dissolved 555 mg CaCl2 in 50mL DIwater
- vacuum filtered into 50mL tube
Made ampicillin stock solution:
-1 g ampicillin -5mL DIwater -ethanol
- filtered by syringe into 15mL tube
- put into ten 1mL alliquots in 1.5mL tubes
- stored in -20°C in ERB lab
Inoculated E. coli DH5α from frozen stock into 12 mL LB broth
- put in 30°C, 200 rpm shaking, overnight
~1.5 hrs
7/8/2010
Alex, Jennifer and Eric - in ERB
Made LB agar plates w/ ampicillin
-10 g LB broth -7.5 g agar -500 mL DI water
- autoclaved mixture at 121°C for 30 min (sterilize time), following Mike Nelson's protocol
Created protocols for Obtaining Deionized Water in the ERB and ERB Spectrophotometer
- uploaded to Team:Michigan/Protocols section
poured 20 LB-amp plates (large)
- left to solidify in ERB 1230
- 12 plates were used by Ann for biobrick transformations
- 8 plates were stored in 4°C ERB
~3.5 hrs work
7/17/2010
Alex & Jennifer - in ERB
Yesterday, Marcus stored newly obtained strains in 4°C in ERB
- W3110 w/ plasmids pTC6 & pET-GFP
- AI-2 reporter (E. coli, amp and kan-resistant)
- MDAI2 w/ plasmids pCT6 & pET-GFP
- AI-2 reporter & LuxS null mutant (E. coli, does not produce AI-2, amp and kan-resistant)
Removed these strains from 4°C -- they are stored in soft agar stab cultures Made one streak plate on LB+amp for each strain from stabs
- placed in 374°C (rm. 1239)
Made one 2mL LB+amp broth cultures in a 15mL tube for each strain from stabs
- placed in 30°C, 200 rpm shaking (rm. 1230)
Placed agar stabs and LB+amp broth in 4°C
~1.5 hrs work
Created protocol/plan for experiment. Testing AI-2 Response
7/19/2010
Alex and Marcus
7/17 plates are no good -- they needed kanamycin
- plates were discarded
- broth cultures had kanamycin added a day later -- too late to be cryostored, but they were transfered to ERB 4°C
Made LB agar
-20 g powder/500 mL DI water
- autoclaved 30 min (sterilize time) 255°C
- poured 4 LB plates
- poured 6 LB+amp plates (100 μg/mL ampicillin)
- poured 10 LB+amp+kan plates (50 μg/mL kanamycin)
- all plates left to cool in ERB 1230
Obtained a rotor for 1.5mL tubes for the centrifuge from Rodger Pinto
- placed in centrifuge in cold room ERB 1224
Obtained LuxS (strain JW2662-1) & MarC (for Jeremy Minty) null mutant E. coli strains from [http://cgsc.biology.yale.edu/ CGSC]
- retrieved from Lin 4°C
- made one spread plate on LB for each strain following Culturing CGSC Strains protocol
- placed both plates in ERB 37°C
Transfered iGEM cryobox from Lin Lab -20°C to ERB -20°C
~6 hrs
7/21/2010
Alex, Eric and Jeremy
{Yesterday, Marcus made broths for E. coli JW2662-1 (LuxS-) (from spread plate), E. coli JW1522-1 (MarC- for Jeremy Minty) (from spread plate), E. coli W3110 (from stab), E. coli MDAI2 (from stab), P. putida for Oil Sands and P. fluorescens for Oil Sands
- 2 mL LB each culture}
Crystored all six strains following Making frozen stocks protocol
- stored in iGEM box -- -80°C Lin Lab
Plated LuxS- and MarC- mutants and Pseudomonas strains on LB Plated W3110 and MDAI2 on LB+amp+kan
- placed all six plates in 37°C ERB 1239
Stored remaining clean LB+amp (6) and LB+amp+kan (8) plates from 7/19 in 4°C ERB 1239
~1.5 hrs
7/22/2010
Alex
During Lab Committee meeting -- Moved the six plates from yesterday from 37°C to 4°C
Uploaded Protocol: Culturing CGSC Strains
7/23/2010
Alex
Updated Strain Database
7/25/2010
Alex and Eric
Made 250 mL LB broth in each of two 500mL flasks
-5 g LB broth -250 mL DI water
- autoclaved 30 min (sterilize time) ~255°C
Autovclaved 250 mL DI water in each of two 500mL flasks
- 30 min sterilize time, ~255°C
Made a 2mL culture in 15mL tube of W3110
-2 mL LB from 4°C -2 μL 100mg/mL ampicillin (final conc. 100μg/ml) -2 μL 50mg/mL kanamycin (final conc. 50μg/mL)
- incubated overnight, 30°C, 200 rpm shaking -- 1pm
Left stuff in autoclave
7/26/2010
Alex and Marcus
{yesterday, Josh and Charlie removed stuff from the autoclave}
Analyzed yesterday's W3110 culture on Lin microplate reader
- endpoint - cuvette - Ex 450nm, Em 520nm, cutoff 515nm (GFP settings)
- blank: 91.591 RFU
- W3110: 117.47 RFU
- fixed Ex 450nm - Em spectrum scan - cuvette
- W3110 - Em peak = 520nm (green - good)
- fixed Em 520nm - Ex spec scan - cuvette
- W3110 - Ex peak = 370nm (wtf) - still good emission at Ex 450nm
- fixed Ex 370 - Em spec scan - cuvette
- W3110 - Em peak = 450nm (wtf)
- endpoint - cuvette - Ex 370nm - Em 450nm
- W3110: 1054.9 RFU
- blank: 1934.6 RFU
- This is some kind of background - very high readings, but the blank is higher than the sample - maybe the GFP in the sample is reabsorbing at 450nm?? At any rate, there appears to be GFP, as expected, and we can probably use Ex 450nm and Em 520nm to detect it.
Autoclaved two 500mL flasks to be sterile containers, each with ~150 mL DI water inside
- 30 min sterilization (55min total), ~250°C
Alex
Learned how to use Epifluorescence Microscope in HHDow (2nd floor) from Alissa
- Viewed yesterday's W3110 culture for GFP
- Unfortunately, no fluorescence could be detected. However, GFP was presumably detected using the microplate reader, so maybe it was just at a very low level. Maybe, the culture was too old (~24 hrs). We will try again tomorrow with a 16-hr culture.
- Uploaded Epifluorescence Microscope Usage protocol
Removed glassware from the autoclave --> ERB 1230
Made broth cultures for W3110 and MDAI2
- each 5 mL LB + 50μg/mL kan + 100μg/mL amp in a 15mL tube (added 5 μL each of a 1000X stock of each antibiotic)
- incubated 30°C, 200 rpm shaking -- 6:30 pm
7/27/2010
Alex, Eric and Marcus
Checked OD600 of yesterday's cultures in Lin Lab spectrometer
- W3110: 0.873
- MDAI2: 0.855
Need to start culture with OD .02
- .02 / .873 = 2.29%
- .02 / .855 = 2.34%
Obtained 40% glucose solution from Ann - Lin Lab
- need .8% glc final conc.
- .8/ 40 = 2%
Started 100mL culture of W3110 in 500mL flask
-2.29 mL W3110 culture -2 mL 40% glucose -95.7 mL LB
- 100 - 2.29 - 2 = 95.71
Started 100mL culture of MDAI2 in 500mL flask
-2.34 mL MDAI2 culture -2 mL 40% glucose -95.7 mL LB
- 100 - 2.34 - 2 = 95.66
- placed both cultures in 37°C, 225 rpm shaking - ERB 1230, 11:50am
Marcus and Eric
Alex and Marcus
Checked remaining MDAI2 and W3110 cultures on fluroescence microscope in LSI 6th floor
- both seem to fluoresce at GFP equally
- This is probably not right; MDAI2 should not have GFP. Will check on a fluorospectrometer tonight
Alex
Read W3110 & MDAI2 cultures on microplate reader in Xi Lab - SPH
- Ex 485nm, Em 545nm (closest to GFP possible on this reader)
- W3110 = 6
- MDAI2 = 5
- Many Acinetobacter controls were run - all at 0 or 1
- This is bad -- MDAI2 is clearly producing GFP, meaning it produces AI-2. Need to contact Tsao authors.
7/28/2010
Alex
Made 40 mL of 0.1% crystal violet solution in Xi Lab - SPH
- for Ann; will deliver tomorrow
7/30/2010
Alex and Marcus - w/ Charlie and Prae
Performed transformation of 5 parts following Transformation-electroporation protocol -- starting from reading OD of overnight. (Ann started the previous steps.)
- read OD of overnights (in Lin Lab)
- JW2662-1 (LuxS-): 1.138 -- bad
- DH5α: 0.710 -- good
- made a 1:2 dilution of JW2662-1:LB (48 mL total)
- placed in 30°C, 200 rpm shaking, 30 min
- removed and read OD
- JW2662-1: 0.657 -- good
- Transfered entire 47 mL of each culture to a 50mL tube
Continued with protocol...
- following spins and washes, read OD again:
- JW2662-1: 0.42 -- good
- DH5α: 0.87 -- good
- electroporated:
- into DH5α:
- pBAD (for Ann)
- INP-GFP (for virus group)
- INP-linker (for virus group)
- OmpA-GFP (for virus group)
- into JW2662-1
- pLsrA-YFP (two sources: L14 and 008)
- time constants:
- OmpA-GFP: 2.8
- INP-GFP: 2.8
- INP-linker: 3.2
- pLsrA-YFP (008): 5.6
- pLsrA-YFP (14L): 5.6
- pBAD: 4.8
- neg. control: 5.0
- incubated all 30°C -- 4:30pm to 7:30pm - 3 hrs
- into DH5α:
- plated on:
- OmpA-GFP: LB+amp
- INP-GFP: LB+amp
- INP-linker: LB+amp
- pLsrA-YFP (both): LB+amp+kan
- pBAD: LB+kan+IPTG
- control: LB+amp AND LB+kan+IPTG
- placed all plates in 37°C
- excess electroporation culture stored in 4°C
8/2/2010
Alex and Eric
Read overnight cultures on Lin Lab spectrometer
- OD600 (endpoint)
- LB: 1.156
- W3110: 1.100
- MDAI2: 1.156
- GFP fluorescence (Ex 450nm, Em 520nm; endpoint)
- LB: 90.291 (RFU)
- W3110: 190.59
- MDAI2: 180.86
- Em 520nm, Ex sweep
- LB: peak 370nm (some kind of background)
- W3110: peak 370nm, kinda bimodal at 440nm (GFP)
- MDAI2: peak 370nm, bimodal at 430nm (GFP)
- Ex 450nm, Em sweep
- LB: peak 515nm......wtf
- W3110: peak ~525nm - ok, GFP
- MDAI2: peak ~525nm - GFP
- Ex 440, Em sweep
- W3110: peak ~520nm
- Ex 440nm, Em 520nm (endpoint)
- LB: 117.61
- W3110: 204.86
- MDAI2: 198.34
OK, idk if these have GFP or what. At any rate, if they do, the MDAI2 is too close to W3110. We'll probably ahve to work with the YFP biobrick instead.
Got C. vulgaris from Bobby Levine
- ~200 mL
- strain 258, from flask 085
- spun 4200 rpm for 10 min
- filtered according to
- stored -20°C ERB 1239
Alex and Marcus
Made broth cultures in 15mL tubes from plates
- 3 mL LB + LuxS-
- 3 mL LB + 100μg/mL amp + [DH5α + pLsr-YFP]
- 3 mL lB + 100μg/mL amp + 50μg/mL kan + [LuxS- + pLsr-YFP]
- placed all in 30°C, 200 rpm shaking
- moved plates back to 4°C
Autoclaved waste flasks
- 55 min (30 sterilize)
- 255°C
8/4/2010
Alex and Marcus
Made broths for experiment tomorrow
- 2 mL LB + 100μg/mL amp + 50μg/mL kan in each of four 15mL tubes - from yesterday's broth cultures
- MDAI2
- LuxS-+pLsrA-YFP
- 2 mL LB + 50μg/mL kan and LuxS- in a 15mL tube - from 7/19 spread plate
- 2 mL LB + 100μg/mL amp and DH5α+pLsrA-YFP - from 7/13 streak plate
- Incubated all 30°C, 200 rpm shaking - 7:20pm
Made stocks of LB + 50μg/mL kan (25 mL) and of LB + 100μg/mL amp + 50μg/mL kan (50 mL)
- each in a 50mL tube
- stored in 4°C
Updated QS experimental protocol
8/5/2010
Alex
Continued testing AI-2 response of yesterday's cultures, following Lsr Circuit Test Protocols
- Obtained cultures of MDAI2 and LuxS- from ERB 1230 at 11:30am (16hrs)
- obtained supernatants:
- MDAI2 (4 hr)
- W3110 (4 hr)
- C. vulgaris (one aliquot)
- moved all to Xi Lab, SPH -- cultures to 4°C
Followed protocol
- OD & GFP/YFP readings (OD600, Ex485/Em545): here
- Similar results with non-GFP strains from Xi Lab. So basically, there is no significant fluorescence in any of these - not even the "positive control" DH5α with pLsrA-YFP. Hopefully this exp works, or it hopefully it will with the new +cntl strain we hope to have soon.
Continued protocol...
- moved plate to RT (bench)
Continued Protocol
- combined pellets & respective supernatants
- plate template:
| well | pellet | supernatant |
|---|---|---|
| A1 | MDAI2 | LB |
| A2 | MDAI2 | MDAI2 |
| A3 | MDAI2 | W3110 |
| A4 | MDAI2 | C. vulgaris |
| B1 | LuxS-+pLsrA-YFP | LB |
| B2 | LuxS-+pLsrA-YFP | MDAI2 |
| B3 | LuxS-+pLsrA-YFP | W3110 |
| B4 | LuxS-+pLsrA-YFP | C. vulgaris |
| D1 | [blank] | LB |
| D2 | [blank] | MDAI2 |
| D3 | [blank] | W3110 |
| D4 | [blank] | C. vulgaris |
- Covered plate w/ gas-permeable membrane
- Started reading in Xi microplate reader
- kinetic; OD600 and Ex485/Em545; 6 hrs at 10min intervals
- 30°C; shaking for 30sec before each reading
Attended Lab Committee meeting
Researched Heidelberg team
uploaded notebook
made presentation for tomorrow
Finished exp by protocol
- processed data and uploaded it
- discarded 15mL tubes
- discarded both 24-well plates
8/19/10
Alex
Acquired new strains (soft agar stabs from Tsao Lab)
- W3110 + pTC6 + pET-GFP
- MDAI2 + pTC6 + pET-GFP
- BL21 + pTC5 + pET-GFP
- produces more AI-2 than W3110
- moved all to ERB 4°C
Made broth cultures of MDAI2 and BL21 from stab cultures
- each 2 mL LB in a 15mL tube
- 37°C, 200 rpm shaking
- placed in incubator at 8pm
Added Quorum Sensing project description to Wiki
--Infekt 04:08, 20 August 2010 (UTC)
8/20/10
Alex
[Earlier, Marcus cryostored BL21 and MDAI2 in Lin -80°C and made a spread plate of new MDAI2 strain.]
Made streak plate of BL21
- placed in 35°C incubator
Made frozen stock of BL21 and MDAI2
- each 500 μL culture + 500 μL 50% glycerol
- stored in ERB -20°C
Updated strain database
8/22/10
Alex
Moved MDAI2 spread plate and BL21 streak plate from 37°C to 4°C
--Infekt 19:21, 22 August 2010 (UTC)
Made BL21 broth culture
- 10 mL LB+100μg/mL amp+50μg/mL kan in a 50mL tube
- 37°C, 200 rpm shaking
- 8:00pm
--Infekt 02:04, 23 August 2010 (UTC)
8/26/10
Alex and Marcus
Made cultures of MDAI2 + pTC6 + pET-GFP (new) and LuxS- + pLsrA-YFP
- each 2 mL LB + 100μg/mL amp + 50μg/mL kan in each of four 15mL tubes (8 tubes total)
- incubated 30°C, 200 rpm shaking, 9:15pm
--Infekt 04:12, 27 August 2010 (UTC)
8/27/10
Alex
Obtained cultures and supernatants from ERB -- 1:15pm (16 hrs); transfered to Xi Lab, SPH
Began Quorum Sensing experiment again, following posted protocol
- Used BL21 supernatant instead of MDAI2
- had to wait until 6:45pm to run exp; cells were overgrown and started to die/pellet
- therefore, took only broth, avoiding pellet, for OD and spin
- spun at 5000rpm instead of 4200
- plate template:
| well | pellet | supernatant |
|---|---|---|
| A1 | MDAI2 | LB |
| A2 | MDAI2 | W3110 |
| A3 | MDAI2 | BL21 |
| A4 | MDAI2 | C. vulgaris |
| B1 | LuxS-+pLsrA-YFP | LB |
| B2 | LuxS-+pLsrA-YFP | W3110 |
| B3 | LuxS-+pLsrA-YFP | BL21 |
| B4 | LuxS-+pLsrA-YFP | C. vulgaris |
| C1 | [blank] | LB |
| C2 | [blank] | MDAI2 |
| C3 | [blank] | W3110 |
| C4 | [blank] | C. vulgaris |
- ran growth curve for 16 hrs (overnight) instead of 6, drawing from Singapore protocol
Returned supernatants to ERB -20°C --Infekt 00:50, 28 August 2010 (UTC)
8/28/10
Alex
Obtained data from QS main experiment
- uploaded here
Only the wells with LB added to pellet had a significant increase in fluorescence over time.
- This doesn't make sense, but seems to replicate earlier results.
It seems that the LB blank well was contaminated slightly. This shouldn't really matter, unless the contamination was also in the other two LB wells, in which case maybe co-culture was what cause the increase in fluorescence. However, OD also only increased significantly for the LB wells, so this is probably why the fluorescence also increased. This does present another experiment idea, however: maybe we can try to culture MDAI2 alone, and then in co-culture with K12 to see if there it a difference in GFP. At any rate, I'm not sure it's feasible to co-culture with algae, so quorum sensing project is probably done.
--Infekt 17:25, 28 August 2010 (UTC)
9/12/10
Alex
Made E. coli K12 broth culture from plate
- 2 mL LB in 15mL tube
- 37°C, 200 rpm shaking - ERB 1230
Uploaded NanR cloning protocol
--Infekt 05:35, 13 September 2010 (UTC)
9/29/10
Alex and John
Dissolved M9 salts to 50X
- All of component A in 1000 mL DIwater
- All of component B in 1000 mL DIwater
Autoclaved both: each split into two 1000mL bottles filled w/ 500mL each (four bottles total)
- 30 min sterilize time
Began biofilm assay following Alex's biofilm assay protocol
Made broth cultures of P. putida LD1, P. fluorescens LD2, and LD1+LD2 coculture from plates, and P. putida KT2440 from Lin Lab frozen stock
- KT2440 inoculated into 2 mL LB in 15mL tube
- others inoculated into 2 mL LB + 100μg/mL amp in 15mL tube
- all incubated in 30°C, 200rpm shaking -- ERB 1230, 8:45pm
Streaked KT2440 on LB plate from frozen
- incubated 37°C -- ERB 1239
Added KT2440 to strain database Mixed M9 A and B solutions and diluted 1/50 in 200 mL sterile DIwater -- in 4 50mL tubes
- each tube 1 mL M9A, 1 mL M9B and 48 mL water
Alex
Uploaded Alex's biofilm assay protocol
Added borax to two of the M9 tubes (100 mL) to buffer at ~pH 9.2
- (.01 mol/L)*(.05 L)*(381.37 g/mol) = 190.7 mg borax/50 mL water
- Tested pH with pH indicator strips
- ~9
- Vacuum-filtered 100 mL w/ steriflips
9/30/10
Alex
Aliquot M9 and buffered M9 into five 15mL tubes each Need 15 mL stocks of each of the following media -- each 15 mL neutral and 15 mL buffered at pH 9:
- M9
- M9 + 0.4% glucose
- M9 + Acros NAs (250mg/L)
- M9 + Sigma Aldrich NAs (250mg/L)
- M9 + both Acros & Sigma NAs (each 250mg/L)
- LB
Added 150 μL 40% glucose to appropriate tubes (.4% final concentration) Aliquot 15 mL LB into each of two 15mL tubes Added NAs to appropriate tubes
- Acros: (.25 mg/mL)*(15mL)/(.91 mg/μL) = 4.12 μL - Sigma: (.25 mg/mL)*(15mL)/(.92 mg/μL) = 4.08 μL
- added 4.1 μL of each NA to each respective tube
Added borax to one tube of LB to buffer at pH9
- (190.7 mg)*(15μL/50μL) = 57.2 mg per 15mL LB
- tested pH with indicator strip
- ~9
Tested pH of neutral M9 with indicator strip
- ~7
Moved KT2440 LB plate from 37°C to 4°C -- ERB 1239
Continued Alex's biofilm assay protocol
- made one 96-well plate for all strains and media at pH7 only
- plate template :
- 100 μL/well: 99 μL medium + 1 μL culture
- covered plate with gas-permeable membrane
- Started 24-hr kinetic reading in Lin Lab microplate reader
- 30°C
- OD600 absorbance
- read every 30 min
Made broth cultures for LD1, LD2 and KT2440 from yesterday's overnight broths
- 2 mL each in a 15mL tube
- 30°C, 200 rpm shaking - 6:20pm
Discarded yesterday's overnight broths
--Infekt 00:57, 1 October 2010 (UTC)
10/1/10
Alex
Moved broth cultures 37°C to 4°C -- 1:30pm (19-hr culture) Prepared biofilm assay plate for all pH9 tests - following Alex's biofilm assay protocol
- plate template:
- 100 μL/well: 99 μL medium + 1 μL culture
- stored plate in Lin Lab 4°C -- 2:20pm
Saved data for curves from ph7 plate Read pH7 plate ODs Performed biofilm assay on ph7 plate Started 24-hr kinetic reading for pH9 plate -- ~midnight
Discarded pH7 plate
Processed pH7 plate data
- uploaded results here
--Infekt 06:55, 2 October 2010 (UTC)
Uploaded pH7 kinetic data here
--Infekt 04:06, 4 October 2010 (UTC)
10/3/10
Alex
Ran CV biofilm assay on pH9 plate, following my protocol
- discarded plate
- uploaded results
- uploaded pH9 kinetic raw data
--Infekt 04:35, 4 October 2010 (UTC)
 "
"