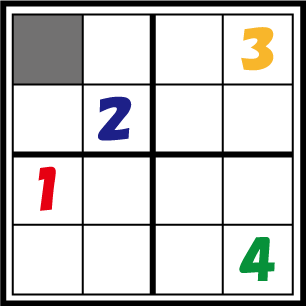Team:UT-Tokyo/Sudoku construct
From 2010.igem.org
(→Combine all systems) |
|||
| (77 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{UT- | + | __NOTOC__{{UT-Tokyo_CSS3}}{{UT-Tokyo_CSS_p}}{{UT-Tokyo_Head}} |
<div id="main_all"> | <div id="main_all"> | ||
= '''Sudoku''' = | = '''Sudoku''' = | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<html> | <html> | ||
| - | < | + | <div> |
| + | <ul id="inpagemenu"> | ||
| + | <li><a href="/Team:UT-Tokyo/Sudoku_abstract" id="abstract">Introduction</a></li> | ||
| + | <li><span>System</span></li> | ||
| + | <li><a href="/Team:UT-Tokyo/Sudoku_modeling" id="modeling">Modeling</a></li> | ||
| + | <li><a href="/Team:UT-Tokyo/Sudoku_experiments" id="experiment">Experiments</a></li> | ||
| + | <li><a href="/Team:UT-Tokyo/Sudoku_perspective" id="perspective">Perspective</a></li> | ||
| + | <li><a href="/Team:UT-Tokyo/Sudoku_reference" id="reference">Reference</a></li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | <div id="clear"></div> | ||
</html> | </html> | ||
| + | == '''System''' == | ||
| + | '''Overall construct''' | ||
| + | [[Image:Sudoku_const_Main_4.png|680px|Sudoku's main construct (We are making more simple version)]] | ||
| + | Here we will explain how our bacteria solve Sudoku. | ||
| - | + | First, we begin by preparing ''E.coli'' corresponding to each cell number. We make these bacteria “decide” what number they should differentiate into. | |
| + | During this process, patial distribution (the position in a 4×4 grid) is temporarily lost. Instead, spatial information is represented by genetic identities assigned to each bacterium. | ||
| + | In fact, the solving process is not performed on a 4×4 grid, but instead in a flask where we mix bacteria possessing all 16 types of genetic spatial identity. | ||
| + | While in this mixture, the puzzle is solved as a result of inter-bacterial interaction. | ||
| + | (We will explain this later in detail.) | ||
| - | |||
| - | |||
| - | + | [[Image:Sudoku_system_1.png|200px|thumb|How do the bacteria “decide” what number they become?]] | |
| - | + | So, how do the bacteria “decide” what number they become? | |
| + | Let’s take a look at the figure. | ||
| + | The number in cell 1-1 has not yet decided. | ||
| + | There are bacteria differentiated as 1 in the same column, 3 in the same row, and 2 in the same block, so the bacteria representing cell 1-1 should differentiate into number 4. | ||
| + | In this way, bacteria which are not assigned a number from the beginning have to decide the number they differentiate into by judging the identity of other bacteria. | ||
| + | The 4C3 leak-switch we suggest enables such a system. | ||
| - | |||
| - | + | Let us explain this switch in detail. | |
| + | This switch outputs the number it did not receive when it receives the other three numbers. | ||
| + | For example, when the switch receives the numbers 1, 2 and 3, it outputs the number 4, and when it receives the numbers 2, 3 and 4, it outputs 1. | ||
| + | The switch must work despite the numbers not being received in a set order, or no matter how many times a number is received. | ||
| + | In other words, it is required that the switch outputs a signal only after receiving three numbers, regardless of the order of reception. | ||
| + | This is the “4C3 leak-switch”. | ||
| - | + | So what is it, in physical terms, what we have been calling “numbers”? | |
| + | The numbers 1 through 4 are represented by 4 different kinds of recombinases. | ||
| + | Bacteria communicate with each other by transmitting RNA encoding these recombinases, | ||
| + | which are packaged in virus vectors. | ||
| + | Look at the figure again. | ||
| + | This communication system must be one that only conveys information between bacteria possessing identities of the same row, column or block. | ||
| + | In this figure, bacteria with the cell identity 1-1 need to differentiate to “4”. Therefore they should not be allowed to receive information from bacteria with the cell identity 4-4 prompting others not to differentiate to “4”. | ||
| + | The signal virus system together with the antisense RNA system enables such restricted communication to take place. We will discuss these systems in detail below. | ||
| - | + | The overall systems therefore are the following: | |
| + | #1. [[#4C3 leak-switch|The 4C3 leak-switch]] designates what number bacteria differentiate into. | ||
| + | #2. [[#Genotype Specific Signal Transmitting Virus|The signal virus]] transmits this information to other bacteria. | ||
| + | #3. [[#Antisense RNA|The antisense RNA system]] restricts exchange of information to that between bacteria possessing identities of the same row, column or block. | ||
| - | + | Now we examine each of these systems in detail. | |
| - | + | == '''4C3 leak-switch''' == | |
| + | [[Image:Sudoku_exp_1_1.png|150px|thumb|4C3 leak switch_1]][[Image:Sudoku_exp_1_2.png|150px|thumb|4C3 leak switch_2]] | ||
| - | + | The 4C3 leak-switch is a switch which receives three number signals and outputs the number signal that it did not receive, regardless of the order of reception. What we call number signals are mRNA encoding homologous recombinases. When three types of mRNA encoding the recombinases enter a bacterium, the switch turns on and transcription of the recombinase that it did not receive begins. The switch is composed of a promoter, the cre protein coding region and four terminators in between. Each terminator is flanked on both sides by the recognition sequence of a homologous recombinase, and a terminator is excised following expression of the corresponding recombinase. Therefore when the ''E.coli'' do not receive a number signal, they possess four terminators; when they receive one number signal, they have three terminators; and so on. | |
| - | + | The 4C3 leak-switch, as the name implies, is a switch that makes use of transcriptional leakage. We assembled the sequence so that when there is more than one terminator, cre protein is not expressed but it is when there is only one. In addition, a recombinase coding sequence flanking the terminator is excised at the same time. Therefore, when the recombinase 1 coding sequence is excised, terminator 1 is also excised. As a result, after a cell receives the number signals 1, 2 and 3, only the recombinase 4 coding sequence remains. Each cre protein expressed in this way irreversibility excises the sequence between the lox sites independently of the other species of cre proteins. In the example above, mRNA coding recombinase 4 corresponding to the answer of this particular puzzle, is transcribed. In conclusion, the 4C3 leak-switch begins transcription of the recombinase that it did not receive begins after receiving three signals. The state in which the bacteria transcribes a recombinase we called differentiated. | |
| + | == '''Genotype Specific Signal Transmitting Virus''' == | ||
| + | [[Image:Sudoku_exp_2.png|200px|thumb|Transcription of MS2 phage & location sequence]] | ||
| - | + | The mRNA coding the homologous recombinases have two additional sequences attached allowing transmission of information to other bacteria. First, they contain a loading sequence to the MS2 phage which enables the mRNA to be attached to the coat protein of MS2 phage. This MS2 phage is the carrier that transfers the number signal to other bacteria. These phage are only synthesized after recombination takes place, therefore preventing undifferentiated bacteria from emitting a number signal precociously. | |
| - | + | Secondly, they contain a location sequence which assigns a cell number to the bacterium. We will explain this in detail later. In conclusion, <br/> | |
| + | 4C3 leak-switch activates<br/> | ||
| + | → transcription of mRNA coding a homologous recombinase<br/> | ||
| + | → production of MS2 phage protein <br/> | ||
| + | → production of phage with the number signal and location sequence attached to it<br/> | ||
| + | In this way, differentiated E.coli produce phage with a number signal and location sequence and conveys this information to other bacteria. | ||
| - | + | == '''Antisense RNA''' == | |
| - | |||
| - | |||
| - | Infection of signal virus. | + | The final component of our system, the antisense RNA system, restricts which bacteria are able to receive which number signals. The phage infect all bacteria, so there needs to be a way for the infected bacteria to shunt away mRNA not targeted to them. This is possible because bacteria representing each cell number transcribes a unique set of antisense RNA. This RNA is antisense to the location sequences included in the mRNA transferred by the phage. Each bacterium has a set of antisense RNA corresponding to the location sequence of bacteria from which are made not to receive a signal from. These antisense RNA attach to the mRNA, preventing transcription, allowing blockage of irrelevant number signals. For instance, bacteria with the identity 1-1 do not want to receive number signals from 1-1, 2-3, 2-4, 3-2, 3-3, 3-4, 4-2, 4-3, 4-4. Therefore, bacteria with identity 1-1 constitutively transcribe RNA antisense to the location sequence of these nine cell numbers and as a result do not receive the number signals from these cells. Thus this system disables bacteria to receive number signals not targeted to them, although the phage infect all bacteria. |
| + | <gallery widths="170"> | ||
| + | Image:Sudoku_exp_3.png|Infection of signal virus. | ||
| + | Image:Sudoku_exp_4_1.png|Selective translational suppression. | ||
| + | Image:Sudoku_const_Antisense_2.png|The construct of selective translational suppression. | ||
| + | </gallery> | ||
| - | |||
| - | + | == '''Movie''' == | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | === | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<html> | <html> | ||
| - | <object width="640" height="385"><param name="movie" value="http://www.youtube.com/v/ | + | <object width="640" height="385"><param name="movie" value="http://www.youtube.com/v/x3IhEUcCV-k?fs=1&hl=ja_JP"></param><param name="allowFullScreen" value="true"></param><param name="allowscriptaccess" value="always"></param><embed src="http://www.youtube.com/v/x3IhEUcCV-k?fs=1&hl=ja_JP" type="application/x-shockwave-flash" allowscriptaccess="always" allowfullscreen="true" width="640" height="385"></embed></object> |
</html> | </html> | ||
| - | == Visualization of results == | + | == '''Visualization of results''' == |
[[Image:Sudoku_const_Result.png|200px|thumb|The construct to visualize results.]] | [[Image:Sudoku_const_Result.png|200px|thumb|The construct to visualize results.]] | ||
| - | + | At this point, you may be wondering how the results are visualized. The differentiation status of bacteria depends on which recombinase they possess. We need to convert this information into a visible form. To do this, we created detection bacteria which will be plated on a 4×4 grid. When these bacteria come in contact with the signal viruses, they are able to receive number signals and fluoresce a different color depending on the signal they receive. Thus, these bacteria recognize the type of number signal (the type of homologous recombinase) and express the corresponding fluorescent protein. As shown, the in plasmids of the detection bacteria a fluorescent protein coding sequence is placed downstream of the terminator, enabling it to be excised simultaneously as the terminator is excised. Therefore, after three signals enter the bacterium and three rounds of excision take place, only one fluorescent protein will remain. The remaining protein is expressed and its color signals to us which number it has differentiated into. For example, if cell 1-1 outputs number 1, only recombinase 1 is transcribed in the detection bacteria, which excises the terminator between the recognition sequences of recombinase 1 and therefore expresses Green Fluorescent Protein. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | == '''Combine all systems''' == | ||
| + | Now that we have discussed each component independently, let us recapitulate the flow of the entire process. | ||
| + | In Sudoku, there are cells which are filled from the beginning and ones which are not. To create a puzzle like the one represented above, bacteria in cell 4-4 need to differentiate into state 4. Therefore we need to infect the undifferentiated bacteria in cell 4-4 with phage possessing number signals 1, 2 and 3 in advance. In this way, if we create undifferentiated bacteria corresponding to each cell and phage possessing the relevant number signals in advance, we can create various “problems” of Sudoku. We place 16 types of bacteria, corresponding to each of the 16 cells, some of them differentiated from the beginning, into a flask to cultivate. During cultivation, the process of transferring number signals from differentiated bacteria to undifferentiated bacteria is repeated and eventually the number all bacteria differentiate into is decided. When this happens, the supernatant of the culture contains phage that possess information on the answer to the puzzle. We extract these phage and spread them on the plate housing detection bacteria. The detection bacteria express a fluorescent protein depending on the number signal they receive, allowing visualization of the answer to the problem. | ||
</div> | </div> | ||
| - | {{UT- | + | {{UT-Tokyo_Foot}} |
Latest revision as of 03:55, 28 October 2010


Sudoku
System
Overall construct
Here we will explain how our bacteria solve Sudoku.
First, we begin by preparing E.coli corresponding to each cell number. We make these bacteria “decide” what number they should differentiate into. During this process, patial distribution (the position in a 4×4 grid) is temporarily lost. Instead, spatial information is represented by genetic identities assigned to each bacterium. In fact, the solving process is not performed on a 4×4 grid, but instead in a flask where we mix bacteria possessing all 16 types of genetic spatial identity. While in this mixture, the puzzle is solved as a result of inter-bacterial interaction. (We will explain this later in detail.)
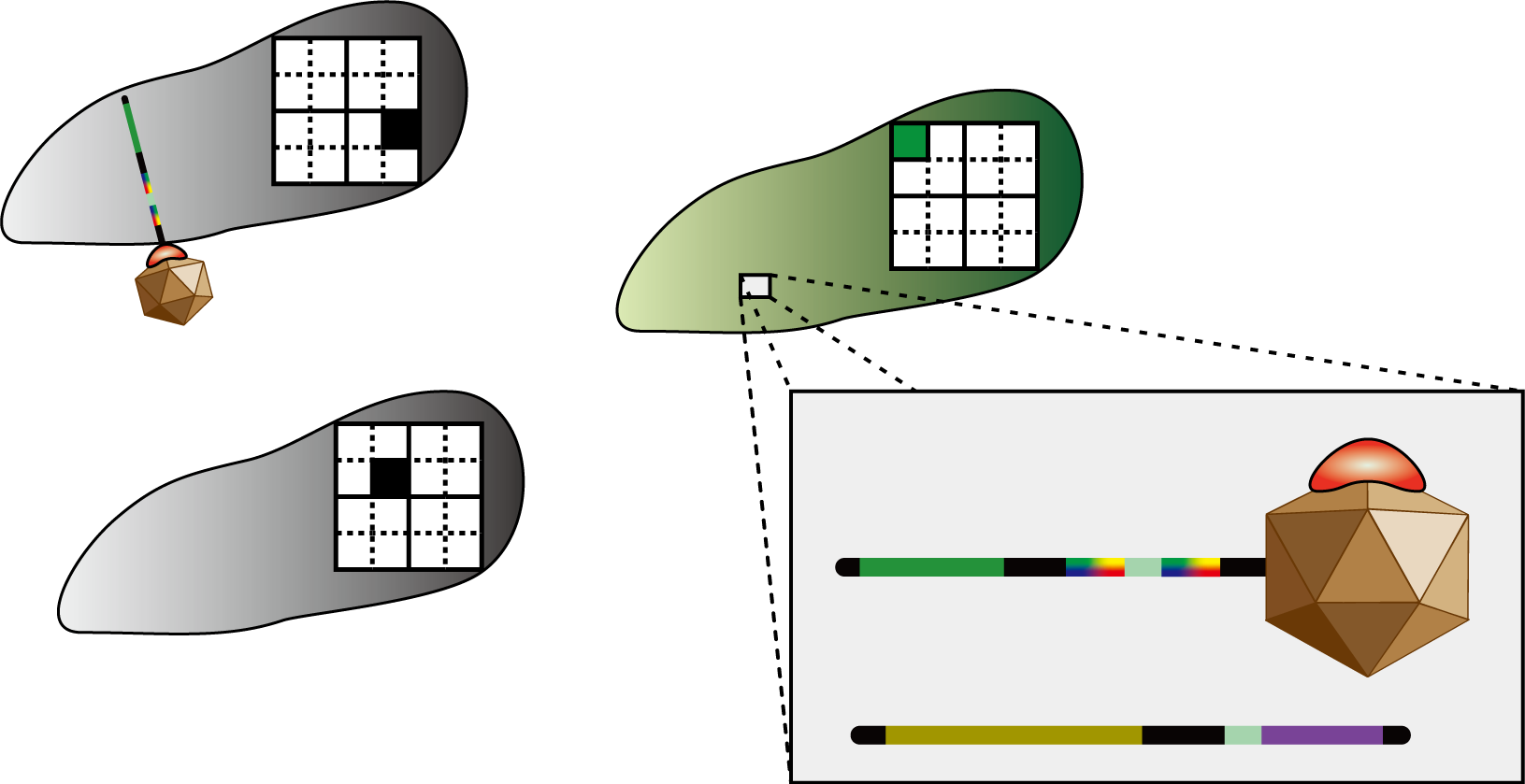
So, how do the bacteria “decide” what number they become? Let’s take a look at the figure. The number in cell 1-1 has not yet decided. There are bacteria differentiated as 1 in the same column, 3 in the same row, and 2 in the same block, so the bacteria representing cell 1-1 should differentiate into number 4. In this way, bacteria which are not assigned a number from the beginning have to decide the number they differentiate into by judging the identity of other bacteria. The 4C3 leak-switch we suggest enables such a system.
Let us explain this switch in detail.
This switch outputs the number it did not receive when it receives the other three numbers.
For example, when the switch receives the numbers 1, 2 and 3, it outputs the number 4, and when it receives the numbers 2, 3 and 4, it outputs 1.
The switch must work despite the numbers not being received in a set order, or no matter how many times a number is received.
In other words, it is required that the switch outputs a signal only after receiving three numbers, regardless of the order of reception.
This is the “4C3 leak-switch”.
So what is it, in physical terms, what we have been calling “numbers”? The numbers 1 through 4 are represented by 4 different kinds of recombinases. Bacteria communicate with each other by transmitting RNA encoding these recombinases, which are packaged in virus vectors. Look at the figure again. This communication system must be one that only conveys information between bacteria possessing identities of the same row, column or block. In this figure, bacteria with the cell identity 1-1 need to differentiate to “4”. Therefore they should not be allowed to receive information from bacteria with the cell identity 4-4 prompting others not to differentiate to “4”. The signal virus system together with the antisense RNA system enables such restricted communication to take place. We will discuss these systems in detail below.
The overall systems therefore are the following:
- 1. The 4C3 leak-switch designates what number bacteria differentiate into.
- 2. The signal virus transmits this information to other bacteria.
- 3. The antisense RNA system restricts exchange of information to that between bacteria possessing identities of the same row, column or block.
Now we examine each of these systems in detail.
4C3 leak-switch
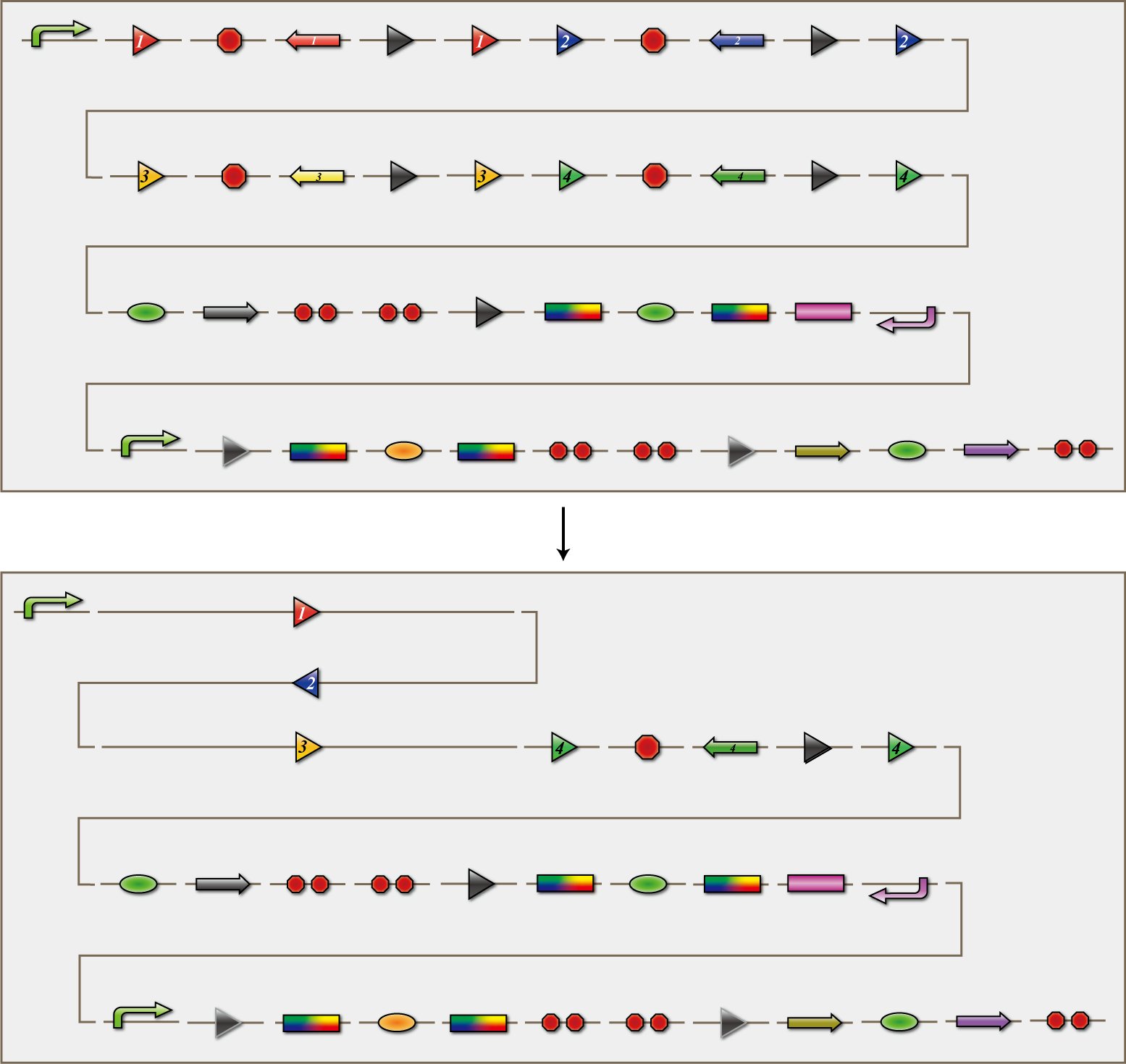
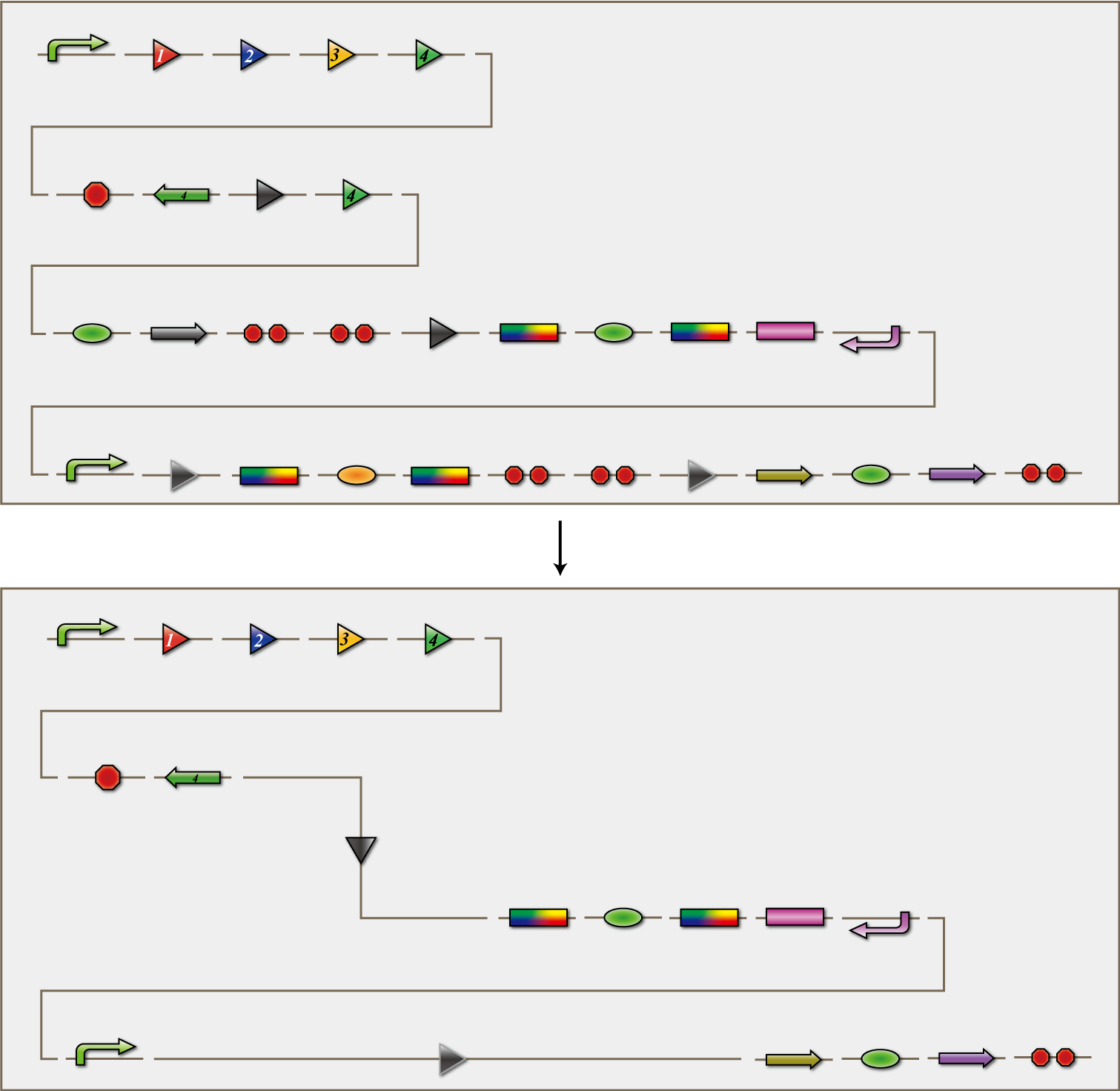
The 4C3 leak-switch is a switch which receives three number signals and outputs the number signal that it did not receive, regardless of the order of reception. What we call number signals are mRNA encoding homologous recombinases. When three types of mRNA encoding the recombinases enter a bacterium, the switch turns on and transcription of the recombinase that it did not receive begins. The switch is composed of a promoter, the cre protein coding region and four terminators in between. Each terminator is flanked on both sides by the recognition sequence of a homologous recombinase, and a terminator is excised following expression of the corresponding recombinase. Therefore when the E.coli do not receive a number signal, they possess four terminators; when they receive one number signal, they have three terminators; and so on.
The 4C3 leak-switch, as the name implies, is a switch that makes use of transcriptional leakage. We assembled the sequence so that when there is more than one terminator, cre protein is not expressed but it is when there is only one. In addition, a recombinase coding sequence flanking the terminator is excised at the same time. Therefore, when the recombinase 1 coding sequence is excised, terminator 1 is also excised. As a result, after a cell receives the number signals 1, 2 and 3, only the recombinase 4 coding sequence remains. Each cre protein expressed in this way irreversibility excises the sequence between the lox sites independently of the other species of cre proteins. In the example above, mRNA coding recombinase 4 corresponding to the answer of this particular puzzle, is transcribed. In conclusion, the 4C3 leak-switch begins transcription of the recombinase that it did not receive begins after receiving three signals. The state in which the bacteria transcribes a recombinase we called differentiated.
Genotype Specific Signal Transmitting Virus
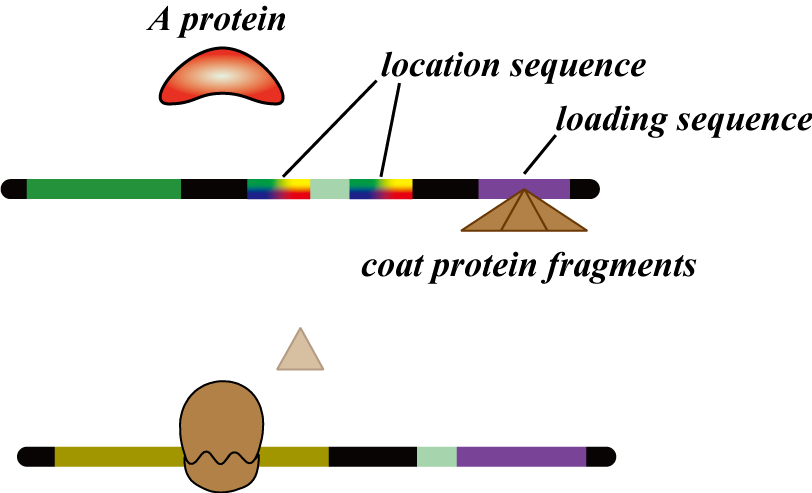
The mRNA coding the homologous recombinases have two additional sequences attached allowing transmission of information to other bacteria. First, they contain a loading sequence to the MS2 phage which enables the mRNA to be attached to the coat protein of MS2 phage. This MS2 phage is the carrier that transfers the number signal to other bacteria. These phage are only synthesized after recombination takes place, therefore preventing undifferentiated bacteria from emitting a number signal precociously.
Secondly, they contain a location sequence which assigns a cell number to the bacterium. We will explain this in detail later. In conclusion,
4C3 leak-switch activates
→ transcription of mRNA coding a homologous recombinase
→ production of MS2 phage protein
→ production of phage with the number signal and location sequence attached to it
In this way, differentiated E.coli produce phage with a number signal and location sequence and conveys this information to other bacteria.
Antisense RNA
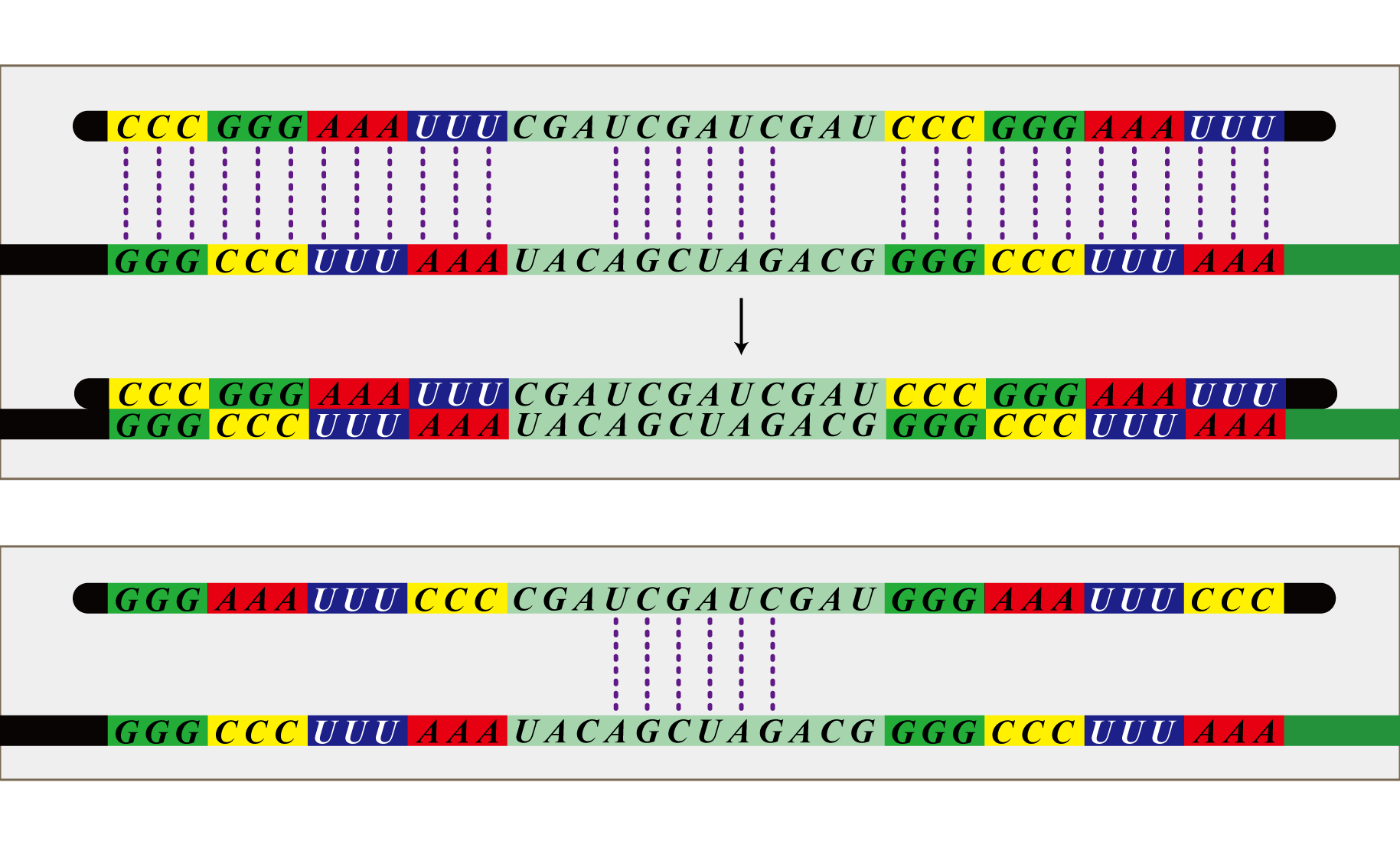
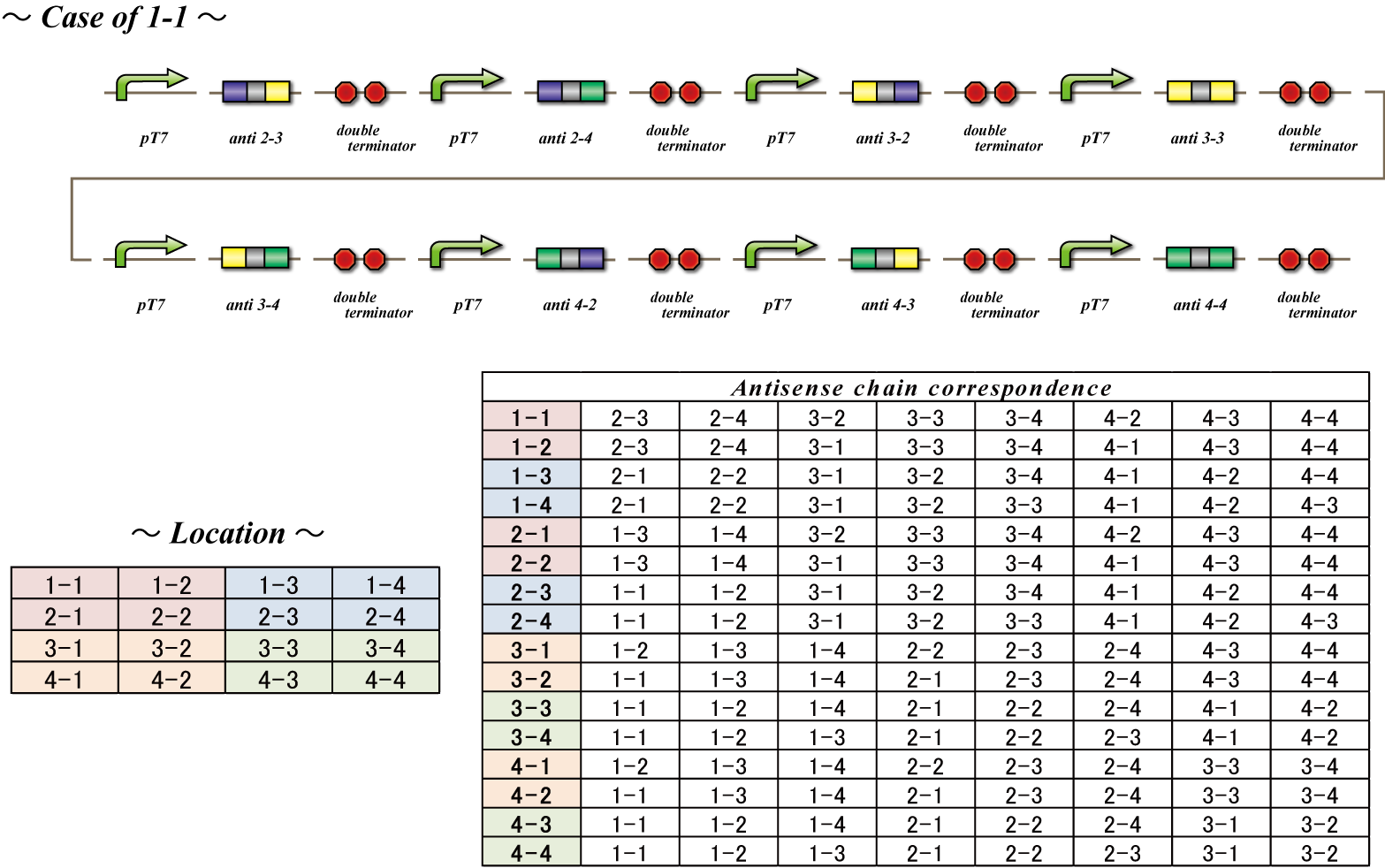
The final component of our system, the antisense RNA system, restricts which bacteria are able to receive which number signals. The phage infect all bacteria, so there needs to be a way for the infected bacteria to shunt away mRNA not targeted to them. This is possible because bacteria representing each cell number transcribes a unique set of antisense RNA. This RNA is antisense to the location sequences included in the mRNA transferred by the phage. Each bacterium has a set of antisense RNA corresponding to the location sequence of bacteria from which are made not to receive a signal from. These antisense RNA attach to the mRNA, preventing transcription, allowing blockage of irrelevant number signals. For instance, bacteria with the identity 1-1 do not want to receive number signals from 1-1, 2-3, 2-4, 3-2, 3-3, 3-4, 4-2, 4-3, 4-4. Therefore, bacteria with identity 1-1 constitutively transcribe RNA antisense to the location sequence of these nine cell numbers and as a result do not receive the number signals from these cells. Thus this system disables bacteria to receive number signals not targeted to them, although the phage infect all bacteria.
Movie
Visualization of results
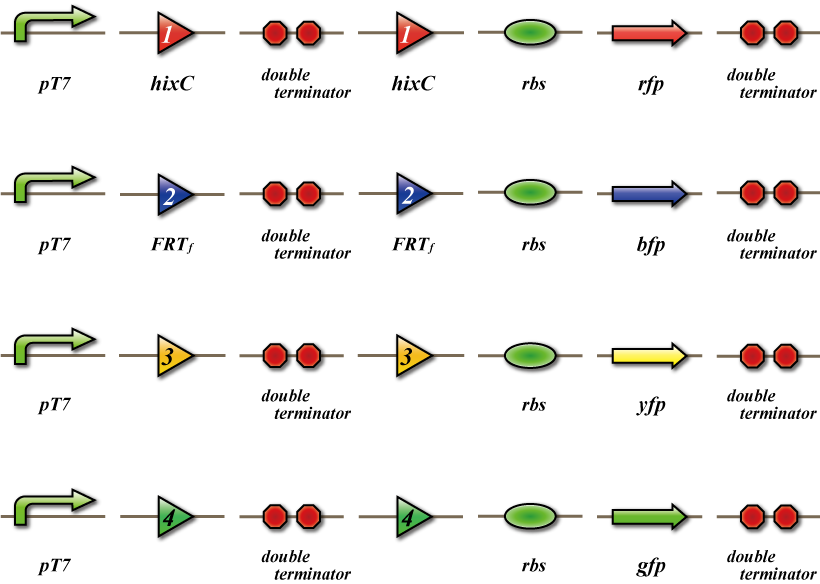
At this point, you may be wondering how the results are visualized. The differentiation status of bacteria depends on which recombinase they possess. We need to convert this information into a visible form. To do this, we created detection bacteria which will be plated on a 4×4 grid. When these bacteria come in contact with the signal viruses, they are able to receive number signals and fluoresce a different color depending on the signal they receive. Thus, these bacteria recognize the type of number signal (the type of homologous recombinase) and express the corresponding fluorescent protein. As shown, the in plasmids of the detection bacteria a fluorescent protein coding sequence is placed downstream of the terminator, enabling it to be excised simultaneously as the terminator is excised. Therefore, after three signals enter the bacterium and three rounds of excision take place, only one fluorescent protein will remain. The remaining protein is expressed and its color signals to us which number it has differentiated into. For example, if cell 1-1 outputs number 1, only recombinase 1 is transcribed in the detection bacteria, which excises the terminator between the recognition sequences of recombinase 1 and therefore expresses Green Fluorescent Protein.
Combine all systems
Now that we have discussed each component independently, let us recapitulate the flow of the entire process. In Sudoku, there are cells which are filled from the beginning and ones which are not. To create a puzzle like the one represented above, bacteria in cell 4-4 need to differentiate into state 4. Therefore we need to infect the undifferentiated bacteria in cell 4-4 with phage possessing number signals 1, 2 and 3 in advance. In this way, if we create undifferentiated bacteria corresponding to each cell and phage possessing the relevant number signals in advance, we can create various “problems” of Sudoku. We place 16 types of bacteria, corresponding to each of the 16 cells, some of them differentiated from the beginning, into a flask to cultivate. During cultivation, the process of transferring number signals from differentiated bacteria to undifferentiated bacteria is repeated and eventually the number all bacteria differentiate into is decided. When this happens, the supernatant of the culture contains phage that possess information on the answer to the puzzle. We extract these phage and spread them on the plate housing detection bacteria. The detection bacteria express a fluorescent protein depending on the number signal they receive, allowing visualization of the answer to the problem.
 "
"