Team:UNIPV-Pavia/Calendar/August/settimana4
From 2010.igem.org
(→August, 24th) |
m (→August, 25th) |
||
| (46 intermediate revisions not shown) | |||
| Line 30: | Line 30: | ||
<html><p align="center"><font size="4"><b>AUGUST: WEEK 4</b></font></p></html><hr><br> | <html><p align="center"><font size="4"><b>AUGUST: WEEK 4</b></font></p></html><hr><br> | ||
| + | <html><a name="indice"/></html> | ||
==August, 23rd== | ==August, 23rd== | ||
| Line 37: | Line 38: | ||
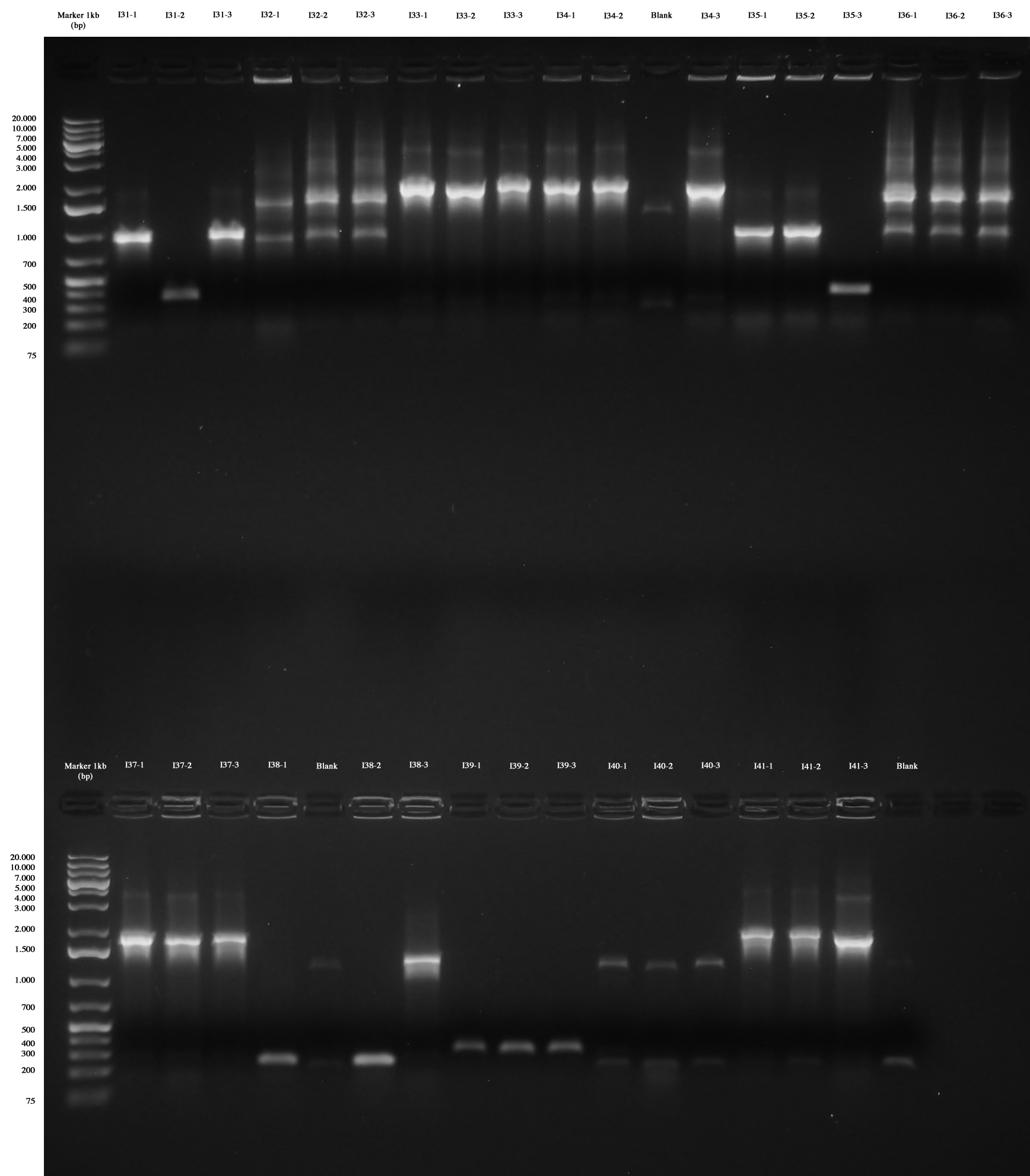
Only I31, I33, I34, I35, I37, I38, I41 are positive without any doubt (we took I31-1, I33-1. I34-1, I35-1, I37-1, I38-3, I41-1), while I39 were all wrong and we weren't sure about I32, I36 and I40 (we made however glycerol stocks for I32-2, I36-2, I40-1). | Only I31, I33, I34, I35, I37, I38, I41 are positive without any doubt (we took I31-1, I33-1. I34-1, I35-1, I37-1, I38-3, I41-1), while I39 were all wrong and we weren't sure about I32, I36 and I40 (we made however glycerol stocks for I32-2, I36-2, I40-1). | ||
We decided to repeat colony PCR for I32, I36, I39 and I40 next day. | We decided to repeat colony PCR for I32, I36, I39 and I40 next day. | ||
| + | |||
| + | |||
| + | MyCrim-9, <partinfo>BBa_K173001</partinfo>, <partinfo>BBa_J23101</partinfo>, <partinfo>pSB4C5</partinfo> were digested E-P and MyCrim-9, Ring were digested HindIII for three hours. | ||
| + | |||
| + | {| border="1" align='center' | ||
| + | | ''Culture'' || ''Kind'' || ''Final reaction volume (ul) '' || ''DNA (ul)'' || ''H20 (ul)'' || ''Enzyme 1 (ul)'' || ''Enzyme 2 (ul)'' || ''Buffer (ul)'' | ||
| + | |- | ||
| + | | MyCrim || Vector || 25 || 6,5 || 14 || 1 EcoRI || 1 PstI || 2,5 H | ||
| + | |- | ||
| + | | MyCrim || Vector/Screening || 25 || 4 || 16,5 || 1 HindIII || 1 HindIII || 2,5 B | ||
| + | |- | ||
| + | | BBa_K173001 || Insert || 25 || 8 || 12,5 || 1 EcoRI || 1 PstI || 2,5 H | ||
| + | |- | ||
| + | | BBa_J23101 || Insert || 25 || 5 || 15,5 || 1 EcoRI || 1 PstI || 2,5 H | ||
| + | |- | ||
| + | | pSB4C5 || Insert || 25 || 5 || 15,5 || 1 EcoRI || 1 PstI || 2,5 H | ||
| + | |- | ||
| + | | Ring || Vector/Screening || 25 || 4 || 16,5 || 1 HindIII || 1 HindIII || 2,5 B | ||
| + | |} | ||
| + | |||
| + | Gel extraction of <partinfo>BBa_I52002</partinfo>(E-P) (insert of pSB4C5), MyCrim (E-P), BBa_K173001 (E-P), MyCrim (HindIII), vector of pSB4C5. | ||
| + | Quantification: | ||
| + | {| border="1" align='center' | ||
| + | | ''Sample'' || ''Quantifications" | ||
| + | |- | ||
| + | | MyCrim (HindIII) || 13 ng/ul | ||
| + | |- | ||
| + | | MyCrim (E-P) || 23,4 ng/ul | ||
| + | |- | ||
| + | | BBa_K173001 (E-P) || 11 ng/ul | ||
| + | |- | ||
| + | | BBa_J23101 (E-P) || 9 ng/ul | ||
| + | |- | ||
| + | | pSB4C5 vector (E-P) || 23 ng/ul | ||
| + | |- | ||
| + | | BBa_I52002 (insert of pSB4C5) (E-P) || 6 ng/ul | ||
| + | |} | ||
| + | |||
| + | These parts were used to perform following ligations: | ||
| + | |||
| + | {| border="1" align='center' | ||
| + | | ''Name'' || ''Vector'' || ''Insert'' | ||
| + | |- | ||
| + | | I42 || MyCrim (E-P) || BBa_K173001 (E-P) | ||
| + | |- | ||
| + | | I43 || MyCrim (E-P) || BBa_J23101 (E-P) | ||
| + | |- | ||
| + | | I44 || MyCrim (E-P) || BBa_I52002 (insert of pSB4C5) (E-P) | ||
| + | |- | ||
| + | | I45 || MyCrim (HindIII) || MyCrim (HindIII) | ||
| + | |} | ||
---- | ---- | ||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==August, 24th== | ==August, 24th== | ||
| - | Only this morning we remembered that I40 is in a commercial vector (pMA), we will screen it next day through an E-P digest (today we made glycerol stocks for other four colonies: I40-4/5/6/7) since it | + | Only this morning we remembered that I40 is in a commercial vector (pMA), we will screen it next day through an E-P digest (today we made glycerol stocks for other four colonies: I40-4/5/6/7) since it's impossible through colony PCR. |
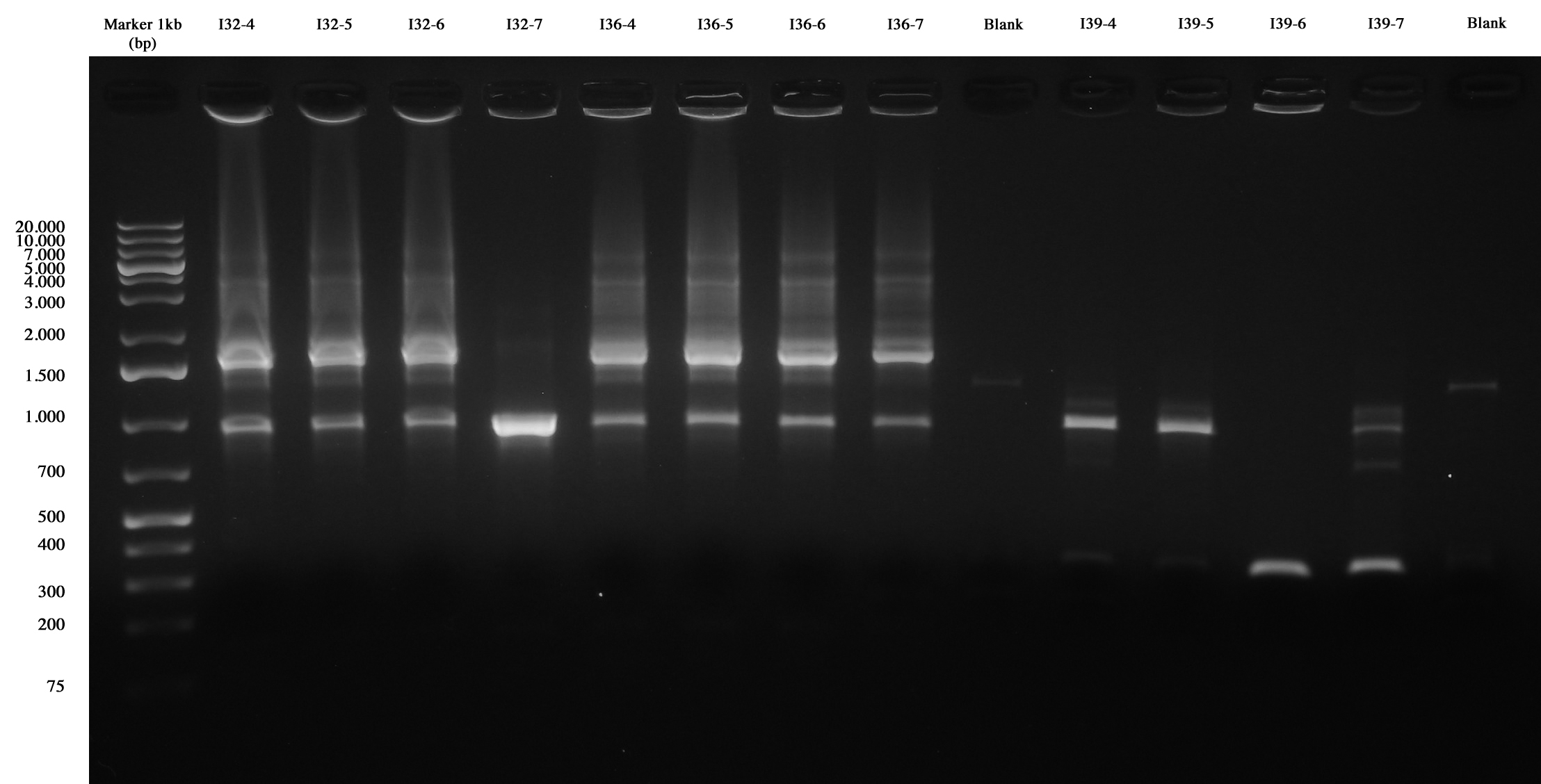
With this method we checked four new colonies of I32 (I32-4/5/6/7), I36 (I36-4/5/6/7) and I39 (I39/4/5/6/7). | With this method we checked four new colonies of I32 (I32-4/5/6/7), I36 (I36-4/5/6/7) and I39 (I39/4/5/6/7). | ||
| Line 53: | Line 106: | ||
Inoculum of: | Inoculum of: | ||
| - | <partinfo>BBa_J23105</partinfo> | + | *<partinfo>BBa_J23105</partinfo> |
| - | <partinfo>BBa_J23106</partinfo> | + | *<partinfo>BBa_J23106</partinfo> |
| - | <partinfo>BBa_J23114</partinfo> | + | *<partinfo>BBa_J23114</partinfo> |
| - | <partinfo>BBa_J23116</partinfo> | + | *<partinfo>BBa_J23116</partinfo> |
for tomorrow MiniPrep. They will be processed with other promoters, for wich we retrieved purified DNA from our freezer. These further parts are: | for tomorrow MiniPrep. They will be processed with other promoters, for wich we retrieved purified DNA from our freezer. These further parts are: | ||
| - | <partinfo>BBa_J23100</partinfo> | + | *<partinfo>BBa_J23100</partinfo> |
| - | <partinfo>BBa_J23101</partinfo> already digested E-P and purified | + | *<partinfo>BBa_J23101</partinfo> already digested E-P and purified |
| - | <partinfo>BBa_J23110</partinfo> | + | *<partinfo>BBa_J23110</partinfo> |
| - | <partinfo>BBa_J23118</partinfo> | + | *<partinfo>BBa_J23118</partinfo> |
| - | <partinfo>pSB4C5</partinfo> already digested E-P and purified | + | *<partinfo>pSB4C5</partinfo> already digested E-P and purified |
These promoters, expressing RFP, will be moved from high copy plasmid <partinfo>pSB1A2</partinfo> to the low copy plasmid <partinfo>pSB4C5</partinfo> and will be tested in both condition in order to establish a strength ranking. | These promoters, expressing RFP, will be moved from high copy plasmid <partinfo>pSB1A2</partinfo> to the low copy plasmid <partinfo>pSB4C5</partinfo> and will be tested in both condition in order to establish a strength ranking. | ||
| + | |||
| + | ---- | ||
| + | Inoculum of PBHR68 BioPlastic producing device and <partinfo>BBa_B0032</partinfo> from glycerol stock in 5ml LB+Amp to check the production of BioPlasic with Sudan Black staining protocol. | ||
| + | |||
| + | Cultures were grown ON at 37°C, 220 rpm. | ||
| + | |||
| + | ---- | ||
| + | I42, I43 ligations were transformed in BW23474 strain and plated on selective LB agar plate with chloramphenicol concentration of 34 ug/ml. Instead I44 was transformed in DB3.1 strain and plated on the same type of plate of the other two transformations. | ||
| + | |||
| + | ---- | ||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==August, 25th== | ==August, 25th== | ||
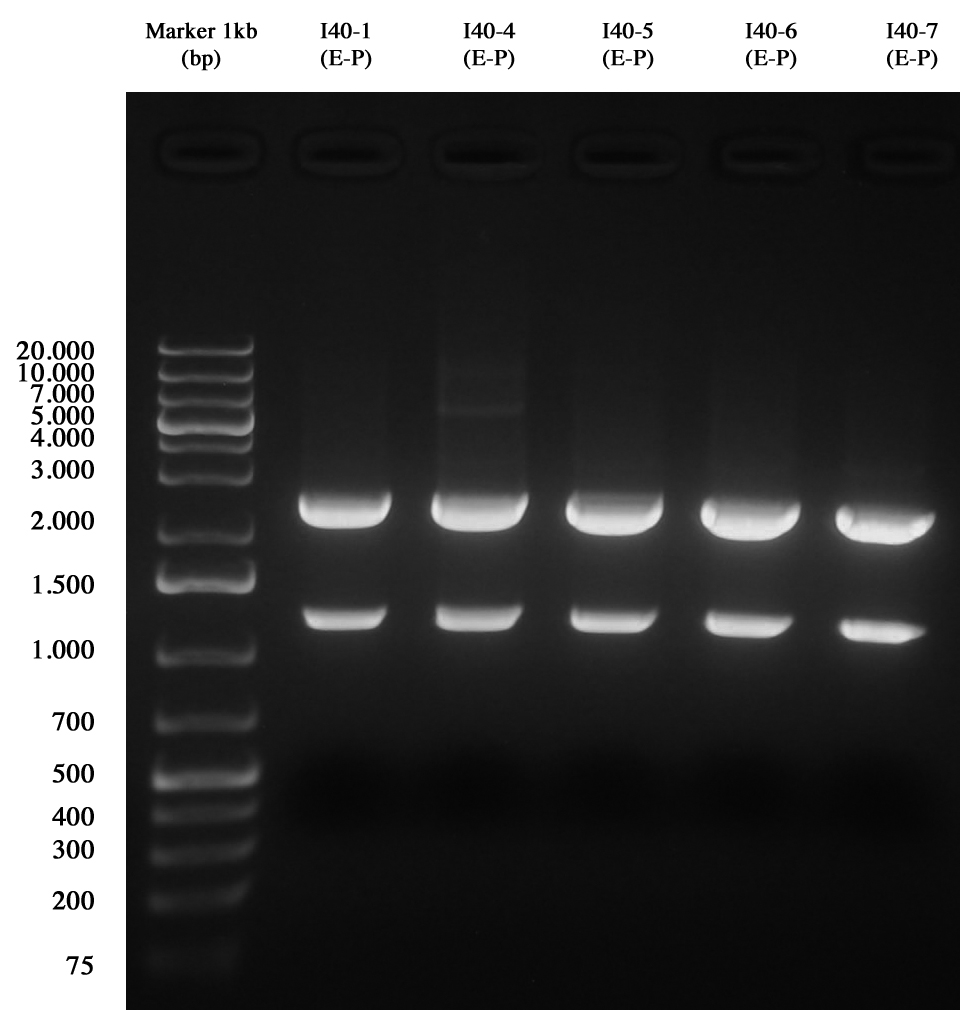
Minipreps of I40-1/4/5/6/7 (to make the screening of ligations) were quantified as follows: | Minipreps of I40-1/4/5/6/7 (to make the screening of ligations) were quantified as follows: | ||
| - | *I40-1: | + | *I40-1: 415,7 ng/ul |
| - | *I40-4: | + | *I40-4: 382,8 ng/ul |
| - | *I40-5: | + | *I40-5: 420 ng/ul |
| - | *I40-6: | + | *I40-6: 487,8 ng/ul |
| - | *I40-7: | + | *I40-7: 479,9 ng/ul |
Samples were digested E-P for three hours | Samples were digested E-P for three hours | ||
| Line 80: | Line 144: | ||
| ''Culture'' || ''Kind'' || ''Final reaction volume (ul) '' || ''DNA (ul)'' || ''H20 (ul)'' || ''Enzyme 1 (ul)'' || ''Enzyme 2 (ul)'' || ''Buffer H (ul)'' | | ''Culture'' || ''Kind'' || ''Final reaction volume (ul) '' || ''DNA (ul)'' || ''H20 (ul)'' || ''Enzyme 1 (ul)'' || ''Enzyme 2 (ul)'' || ''Buffer H (ul)'' | ||
|- | |- | ||
| - | | I40-1 || Insert/Screening || 25 || | + | | I40-1 || Insert/Screening || 25 || 2 || 19,5 || 0,5 EcoRI || 0,5 PstI || 2,5 |
|- | |- | ||
| - | | I40-4 || Insert/Screening || 25 || | + | | I40-4 || Insert/Screening || 25 || 2 || 19,5 || 0,5 EcoRI || 0,5 PstI || 2,5 |
|- | |- | ||
| - | | I40-5 | + | | I40-5 || Insert/Screening || 25 || 2 || 19,5 || 0,5 EcoRI || 0,5 PstI || 2,5 |
|- | |- | ||
| - | | I40-6 | + | | I40-6|| Insert/Screening || 25 || 2 || 19,5 || 0,5 EcoRI || 0,5 PstI || 2,5 |
|- | |- | ||
| - | | I40-7 | + | | I40-7 || Insert/Screening || 25 || 2 || 19,5 || 0,5 EcoRI || 0,5 PstI || 2,5 |
|} | |} | ||
and than gel run to check the length of ligations. | and than gel run to check the length of ligations. | ||
[[Image:UNIPV10_25_8_10_E-PscreeningI40.jpg|thumb|300px|center|Gel run for I40-1/4/5/6/7 digested E-P]] | [[Image:UNIPV10_25_8_10_E-PscreeningI40.jpg|thumb|300px|center|Gel run for I40-1/4/5/6/7 digested E-P]] | ||
| - | As you can see | + | As you can see all samples are positive, so we decided to keep glycerol stock of I40-1. |
---- | ---- | ||
| + | |||
| + | MiniPrep was performed for following cultures, and DNA was quantified as follows: | ||
| + | {| align='center' border='1' | ||
| + | | '''Culture name''' | ||
| + | |- | ||
| + | | <partinfo>BBa_J23105</partinfo> | ||
| + | |- | ||
| + | | <partinfo>BBa_J23106</partinfo> | ||
| + | |- | ||
| + | | <partinfo>BBa_J23114</partinfo> | ||
| + | |- | ||
| + | | <partinfo>BBa_J23116</partinfo> | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | Purified DNA was digested as follows: | ||
| + | |||
| + | |||
| + | Digestion of: | ||
| + | |||
| + | {| border="1" align='center' | ||
| + | | ''Culture'' || ''Kind'' || ''Final reaction volume (ul) '' || ''DNA (ul)'' || ''H20 (ul)'' || ''Enzyme 1'' || ''Enzyme 2'' || ''Buffer H'' | ||
| + | |- | ||
| + | | <partinfo>BBa_J23100</partinfo> || Insert || 25 || 10 || 10,5 || 1 EcoRI || 1 PstI || 2,5 | ||
| + | |- | ||
| + | | <partinfo>BBa_J23105</partinfo> || Insert || 25 || 10 || 10,5 || 1 EcoRI || 1 PstI || 2,5 | ||
| + | |- | ||
| + | | <partinfo>BBa_J23106</partinfo> || Insert || 25 || 10 || 10,5 || 1 EcoRI || 1 PstI || 2,5 | ||
| + | |- | ||
| + | | <partinfo>BBa_J23110</partinfo> || Insert || 25 || 10 || 10,5 || 1 EcoRI || 1 PstI || 2,5 | ||
| + | |- | ||
| + | | <partinfo>BBa_J23114</partinfo> || Insert || 25 || 10 || 10,5 || 1 EcoRI || 1 PstI || 2,5 | ||
| + | |- | ||
| + | | <partinfo>BBa_J23116</partinfo> || Insert || 25 || 10 || 10,5 || 1 EcoRI || 1 PstI || 2,5 | ||
| + | |- | ||
| + | | <partinfo>BBa_J23118</partinfo> || Insert || 25 || 10 || 10,5 || 1 EcoRI || 1 PstI || 2,5 | ||
| + | |||
| + | |} | ||
| + | |||
| + | Ligation of: | ||
| + | |||
| + | * <partinfo>BBa_J23100</partinfo>_4C5=<partinfo>BBa_J23100</partinfo>(E-P)+<partinfo>pSB4C5</partinfo>(E-P) | ||
| + | * <partinfo>BBa_J23101</partinfo>_4C5=<partinfo>BBa_J23101</partinfo>(E-P)+<partinfo>pSB4C5</partinfo>(E-P) | ||
| + | * <partinfo>BBa_J23105</partinfo>_4C5=<partinfo>BBa_J23105</partinfo>(E-P)+<partinfo>pSB4C5</partinfo>(E-P) | ||
| + | * <partinfo>BBa_J23106</partinfo>_4C5=<partinfo>BBa_J23106</partinfo>(E-P)+<partinfo>pSB4C5</partinfo>(E-P) | ||
| + | * <partinfo>BBa_J23110</partinfo>_4C5=<partinfo>BBa_J23110</partinfo>(E-P)+<partinfo>pSB4C5</partinfo>(E-P) | ||
| + | * <partinfo>BBa_J23114</partinfo>_4C5=<partinfo>BBa_J23114</partinfo>(E-P)+<partinfo>pSB4C5</partinfo>(E-P) | ||
| + | * <partinfo>BBa_J23116</partinfo>_4C5=<partinfo>BBa_J23116</partinfo>(E-P)+<partinfo>pSB4C5</partinfo>(E-P) | ||
| + | * <partinfo>BBa_J23118</partinfo>_4C5=<partinfo>BBa_J23118</partinfo>(E-P)+<partinfo>pSB4C5</partinfo>(E-P) | ||
| + | |||
| + | ---- | ||
| + | |||
| + | |||
| + | Preparation of samples for BioPlastic screening: | ||
| + | |||
| + | * Cultures of PBHR68 and <partinfo>BBa_B0032</partinfo> were diluted 1:100 in fresh LB+Amp and werew prepared as follows: | ||
| + | ** <partinfo>BBa_B0032</partinfo> with NOTHING added (negative control) | ||
| + | ** PBHR68 with NOTHING added | ||
| + | ** PBHR68 + 2% glycerol (carbon source for BioPlastic production) | ||
| + | ** PBHR68 + 1mM IPTG (inducer for Plac promoter, expressing BioPlastic enzymes) | ||
| + | ** PBHR68 + 2% glycerol + 1mM IPTG | ||
| + | |||
| + | After 8 hours, Sudan Black staining protocol was performed on 70ul cultures and 5 microscope slides were prepared. The resulting images are shown here: | ||
| + | |||
| + | {|align="center" | ||
| + | |- | ||
| + | |[[Image:UNIPV10_PBHR68_nothing.jpg|thumb|200px|center|PBHR68 with nothing added in the culture, after 8 hours]] || |[[Image:UNIPV10_PBHR68_nothing_2.jpg|thumb|200px|center|PBHR68 with nothing added in the culture, after 8 hours]] | ||
| + | |- | ||
| + | |[[Image:UNIPV10_RBS32_nothing.jpg|thumb|200px|center|<partinfo>BBa_B0032</partinfo> with nothing added in the culture, after 8 hours (negative control)]] || |[[Image:UNIPV10_RBS32_nothing_2.jpg|thumb|200px|center|<partinfo>BBa_B0032</partinfo> with nothing added in the culture, after 8 hours (negative control)]] | ||
| + | |- | ||
| + | |[[Image:UNIPV10_PBHR68_gly.jpg|thumb|200px|center|PBHR68 with 2% glycerol added in the culture, after 8 hours]] || |[[Image:UNIPV10_PBHR68_gly_2.jpg|thumb|200px|center|PBHR68 with 2% glycerol added in the culture, after 8 hours]] | ||
| + | |- | ||
| + | |[[Image:UNIPV10_PBHR68_IPTG.jpg|thumb|200px|center|PBHR68 with 1mM IPTG added in the culture, after 8 hours]] || |[[Image:UNIPV10_PBHR68_IPTG_2.jpg|thumb|200px|center|PBHR68 with 1mM IPTG in the culture, after 8 hours]] | ||
| + | |- | ||
| + | |[[Image:UNIPV10_PBHR68_gly_IPTG.jpg|thumb|200px|center|PBHR68 with 1mM IPTG and 2% glycerol added in the culture, after 8 hours]] || |[[Image:UNIPV10_PBHR68_gly_IPTG_2.jpg|thumb|200px|center|PBHR68 with 1mM IPTG and 2% glycerol added in the culture, after 8 hours]] | ||
| + | |} | ||
| + | |||
| + | Cultures were further incubated for 22 hours at 37°C, 220 rpm. Tomorrow, we will repeat the staining protocol after 30 hours from inoculum to check the time ''E. coli'' takes to produce BioPlastic granules. | ||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==August, 26th== | ==August, 26th== | ||
| + | |||
| + | Transformation of ligations: | ||
| + | |||
| + | {| border='1' align='center' | ||
| + | |'''Ligation name''' || '''Strain''' || '''Resistance''' | ||
| + | |- | ||
| + | |<partinfo>BBa_J23100</partinfo>_4C5 || TOP10 || Cm 12,5 | ||
| + | |- | ||
| + | |<partinfo>BBa_J23101</partinfo>_4C5 || TOP10 || Cm 12,5 | ||
| + | |- | ||
| + | |<partinfo>BBa_J23105</partinfo>_4C5 || TOP10 || Cm 12,5 | ||
| + | |- | ||
| + | |<partinfo>BBa_J23106</partinfo>_4C5 || TOP10 || Cm 12,5 | ||
| + | |- | ||
| + | |<partinfo>BBa_J23110</partinfo>_4C5 || TOP10 || Cm 12,5 | ||
| + | |- | ||
| + | |<partinfo>BBa_J23114</partinfo>_4C5 || TOP10 || Cm 12,5 | ||
| + | |- | ||
| + | |<partinfo>BBa_J23116</partinfo>_4C5 || TOP10 || Cm 12,5 | ||
| + | |- | ||
| + | |<partinfo>BBa_J23118</partinfo>_4C5 || TOP10 || Cm 12,5 | ||
| + | |} | ||
| + | |||
| + | Plates were incubated at 37°C overnight. | ||
| + | |||
| + | Inoculum of self inducible promoters for TECAN test: | ||
| + | |||
| + | {| border='1' | ||
| + | | I7 ||I8 || I9 || I10 | ||
| + | |- | ||
| + | | I12 || I8_4C5 || I15 || I16 | ||
| + | |- | ||
| + | | I17 || I18|| I19 || I27 | ||
| + | |- | ||
| + | | I28 || I30|| <partinfo>BBa_B0032</partinfo> || A2 (<partinfo>BBa_J23101</partinfo>+GFP) | ||
| + | |} | ||
| + | |||
| + | ---- | ||
| + | After 30 hours, Sudan Black staining protocol was performed on 70ul cultures and 5 microscope slides were prepared. The resulting images are shown here: | ||
| + | |||
| + | {|align="center" | ||
| + | |- | ||
| + | |[[Image:UNIPV10_PBHR68_nothing_30.jpg|thumb|200px|center|PBHR68 with nothing added in the culture, after 30 hours]] || |[[Image:UNIPV10_PBHR68_nothing_2_30.jpg|thumb|200px|center|PBHR68 with nothing added in the culture, after 30 hours]] | ||
| + | |- | ||
| + | |[[Image:UNIPV10_RBS32_nothing_30.jpg|thumb|200px|center|<partinfo>BBa_B0032</partinfo> with nothing added in the culture, after 30 hours (negative control)]] || |[[Image:UNIPV10_RBS32_nothing_2_30.jpg|thumb|200px|center|<partinfo>BBa_B0032</partinfo> with nothing added in the culture, after 30 hours (negative control)]] | ||
| + | |- | ||
| + | |[[Image:UNIPV10_PBHR68_gly_30.jpg|thumb|200px|center|PBHR68 with 2% glycerol added in the culture, after 30 hours]] || |[[Image:UNIPV10_PBHR68_gly_2_30.jpg|thumb|200px|center|PBHR68 with 2% glycerol added in the culture, after 30 hours]] | ||
| + | |- | ||
| + | |[[Image:UNIPV10_PBHR68_IPTG_30.jpg|thumb|200px|center|PBHR68 with 1mM IPTG added in the culture, after 30 hours]] || |[[Image:UNIPV10_PBHR68_IPTG_2_30.jpg|thumb|200px|center|PBHR68 with 1mM IPTG in the culture, after 30 hours]] | ||
| + | |- | ||
| + | |[[Image:UNIPV10_PBHR68_gly_IPTG_30.jpg|thumb|200px|center|PBHR68 with 1mM IPTG and 2% glycerol added in the culture, after 30 hours]] || |[[Image:UNIPV10_PBHR68_gly_IPTG_2_30.jpg|thumb|200px|center|PBHR68 with 1mM IPTG and 2% glycerol added in the culture, after 30 hours]] | ||
| + | |} | ||
| + | |||
| + | ---- | ||
| + | Inoculum of | ||
| + | *<partinfo>BBa_J13002</partinfo> | ||
| + | *I33 | ||
| + | *I34 | ||
| + | *I39 | ||
| + | *I41 | ||
| + | in 5 ml LB+Amp | ||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==August, 27th== | ==August, 27th== | ||
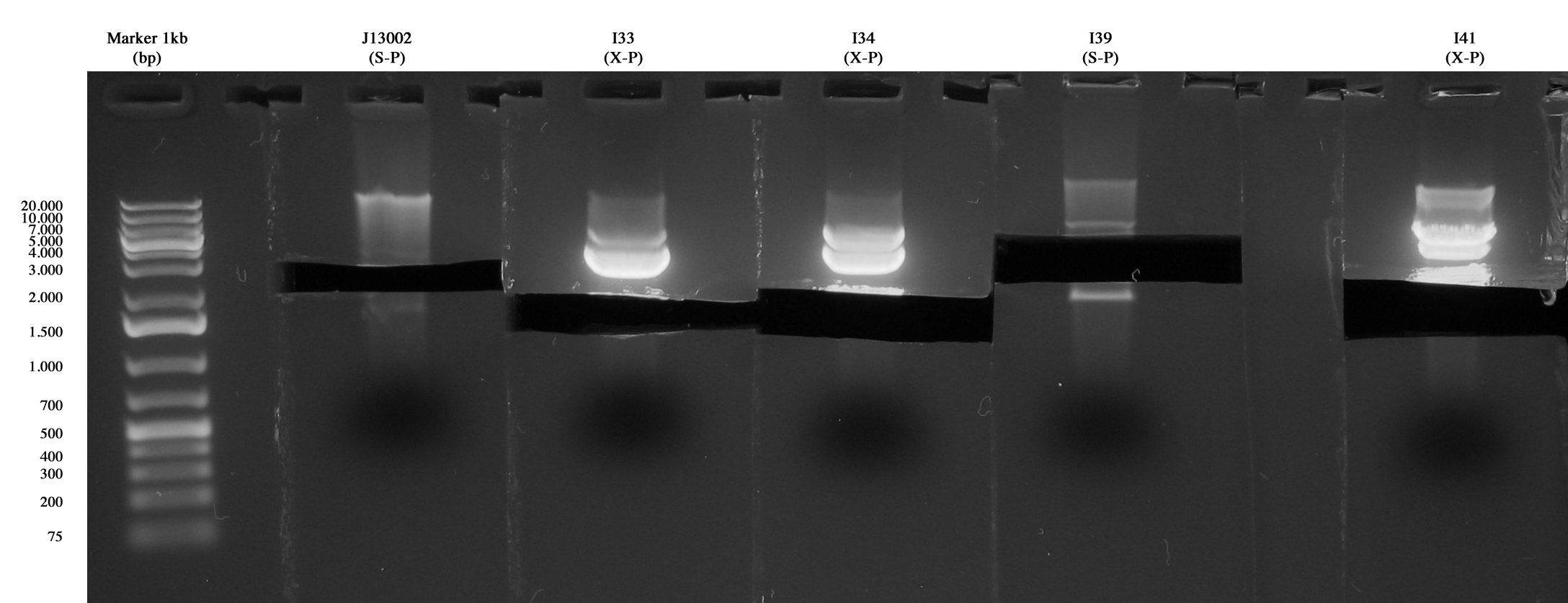
| + | Previously inoculated strains were miniprepped and quantified as follows: | ||
| + | *<partinfo>BBa_J13002</partinfo>: 29,9 ng/ul | ||
| + | *I33: 171,6 ng/ul | ||
| + | *I34: 219,4 ng/ul | ||
| + | *I39: 21,2 ng/ul | ||
| + | *I41: 94,7 ng/ul | ||
| - | + | Digestions (3,5 hours) for ligations: | |
| + | *I47: <partinfo>BBa_J13002</partinfo> (S-P) + I33 (X-P) | ||
| + | *I48: I39 (S-P) + I34 (X-P) | ||
| + | *I49: I39 (S-P) + I41 (X-P) | ||
| - | == | + | {| border="1" align='center' |
| + | | ''Culture'' || ''Kind'' || ''Final reaction volume (ul) '' || ''DNA (ul)'' || ''H20 (ul)'' || ''Enzyme 1 (ul)'' || ''Enzyme 2 (ul)'' || ''Buffer H (ul)'' | ||
| + | |- | ||
| + | | <partinfo>BBa_J13002</partinfo> || Vector || 25 || 21,5 || 0 || 0,5 SpeI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I39 || Vector || 25 || 21,5 || 0 || 0,5 S || 0,5 P || 2,5 | ||
| + | |- | ||
| + | | I33 || Insert || 25 || 10,5 || 11 || 0,5 XbaI || 0,5 P || 2,5 | ||
| + | |- | ||
| + | | I34 || Insert|| 25 || 13,5 || 8 || 0,5 X || 0,5 P || 2,5 | ||
| + | |- | ||
| + | | I41 || Insert || 25 || 2 || 19,5 || 0,5 X || 0,5 P || 2,5 | ||
| + | |} | ||
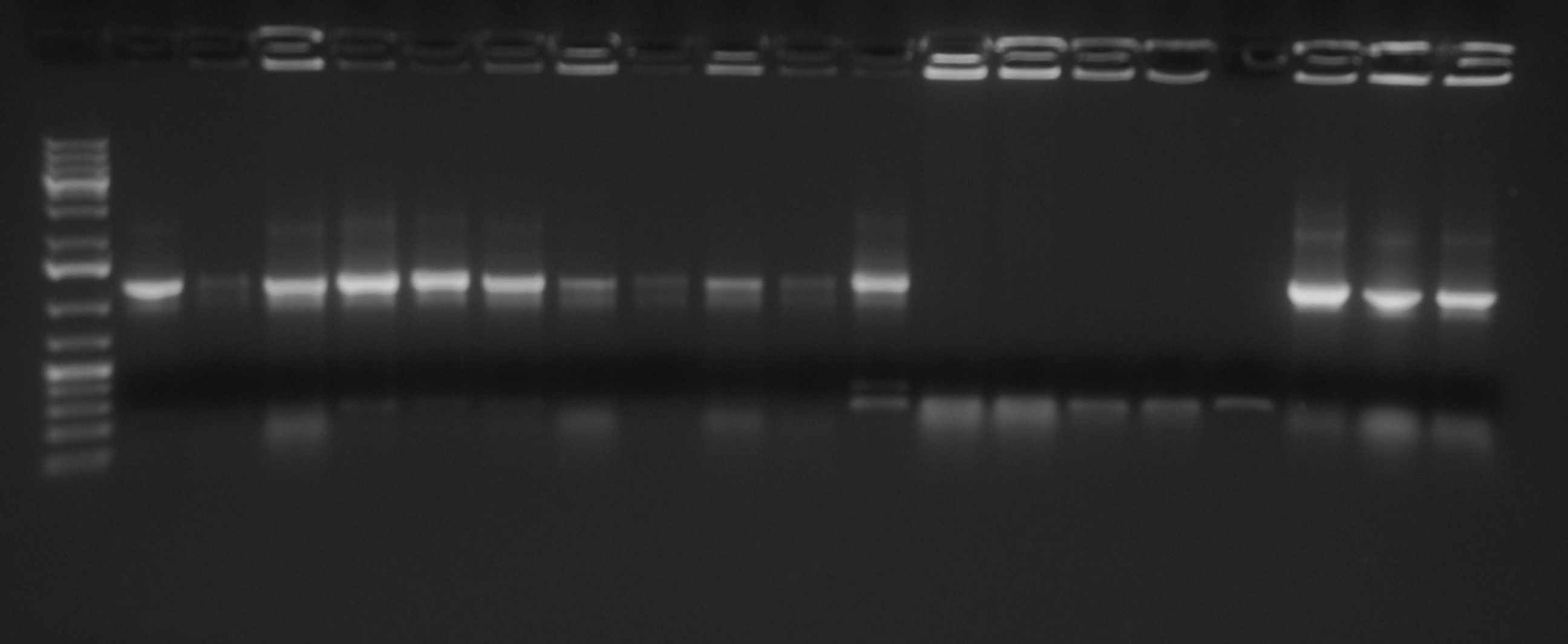
| + | Samples were loaded into a medium gel, run and cut | ||
| + | [[Image:UNIPV10_27_8_10_digest_step2A.jpg|thumb|300px|center|Samples gel run/cut]] | ||
| + | |||
| + | Gel extraction was quantified as follows: | ||
| + | *<partinfo>BBa_J13002</partinfo> (S-P): 19,9 ng/ul | ||
| + | *I33 (X-P): 17,7 ng/ul | ||
| + | *I34 (X-P): 11,1 ng/ul | ||
| + | *I39 (S-P): 4,7 ng/ul | ||
| + | *I41 (X-P): 6,2 ng/ul | ||
| + | |||
| + | Ligation of | ||
| + | *I47: <partinfo>BBa_J13002</partinfo> (S-P) + I33 (X-P) | ||
| + | *I48: I39 (S-P) + I34 (X-P) | ||
| + | *I49: I39 (S-P) + I41 (X-P) | ||
| + | |||
| + | |||
| + | ---- | ||
| + | |||
| + | Plates containing the transformed ligations were all grown. Colony PCR was performed to screen if the length of insert was correct. | ||
| + | |||
| + | {| align='center' border='1' | ||
| + | | '''ligation name'''|| '''number of colonies screened''' | ||
| + | |- | ||
| + | | <partinfo>BBa_J23100</partinfo>_4C5 || 2 colonies | ||
| + | |- | ||
| + | | <partinfo>BBa_J23101</partinfo>_4C5 || 2 colonies | ||
| + | |- | ||
| + | | <partinfo>BBa_J23105</partinfo>_4C5 || 2 colonies | ||
| + | |- | ||
| + | | <partinfo>BBa_J23106</partinfo>_4C5 || 2 colonies | ||
| + | |- | ||
| + | | <partinfo>BBa_J23110</partinfo>_4C5 || 2 colonies | ||
| + | |- | ||
| + | | <partinfo>BBa_J23114</partinfo>_4C5 || 1 colony | ||
| + | |- | ||
| + | | <partinfo>BBa_J23116</partinfo>_4C5 || 2 colonies | ||
| + | |- | ||
| + | | <partinfo>BBa_J23118</partinfo>_4C5 || 2 colonies | ||
| + | |} | ||
| + | |||
| + | |||
| + | [[Image:UNIPV10_screening_promoters_Anserson_low_copy.jpg|thumb|300px|center|PCR screening for Anderson Promoters transferred in low copy plasmid <partinfo>pSB4C5</partinfo>]] | ||
| + | |||
| + | Gel results show that we have the correct clones for <partinfo>BBa_J23100</partinfo>_4C5, <partinfo>BBa_J23101</partinfo>_4C5, <partinfo>BBa_J23105</partinfo>_4C5, <partinfo>BBa_J23106</partinfo>_4C5, <partinfo>BBa_J23110</partinfo>_4C5, <partinfo>BBa_J23114</partinfo>_4C5. Glycerol stocks were prepared for these parts and are stored at -80°C. | ||
| + | |||
| + | <partinfo>BBa_J23116</partinfo>_4C5 and <partinfo>BBa_J23118</partinfo>_4C5 need to be further screened, so we will pick other colonies next week and we will perform a new colony PCR. | ||
| + | ---- | ||
| + | |||
| + | <font size=4>[[Team:UNIPV-Pavia/Material Methods/Measurements/Tecan/test27agosto|Tecan Test]]</font> was performed on prepared samples, after the usual protocol (dilution, medium change and dilution 1:1000). | ||
| + | |||
| + | ---- | ||
| + | Transformation of I29 in ''E. coli'' TOP10 | ||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
| + | |||
| + | ==August, 28th== | ||
| + | Ligations I47, I48, I49 were stored at +4°C. | ||
| + | |||
| + | ---- | ||
| + | I29 plate was stored at +4°C. | ||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
<!-- table previous next week --> | <!-- table previous next week --> | ||
| Line 110: | Line 396: | ||
<tr> | <tr> | ||
<td align="left">[[Team:UNIPV-Pavia/Calendar/August/settimana3| Previous week]]</td> | <td align="left">[[Team:UNIPV-Pavia/Calendar/August/settimana3| Previous week]]</td> | ||
| - | <td align="right">Next week</td> | + | <td align="right">[[Team:UNIPV-Pavia/Calendar/September/settimana1| Next week]]</td> |
</tr> | </tr> | ||
</table> | </table> | ||
Latest revision as of 16:07, 23 October 2010
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"