Team:Stockholm/20 August 2010
From 2010.igem.org
(→Andreas) |
|||
| (11 intermediate revisions not shown) | |||
| Line 124: | Line 124: | ||
==Andreas== | ==Andreas== | ||
| - | |||
| - | |||
| - | + | ===Transformation results=== | |
| + | ''From 19/8 transformations'' | ||
*pEX.RFP: Good yield of red (positive) and white (negative) colonies. | *pEX.RFP: Good yield of red (positive) and white (negative) colonies. | ||
*pSB1K3.SOD⋅His: Very few colonies | *pSB1K3.SOD⋅His: Very few colonies | ||
| Line 135: | Line 134: | ||
====Colony PCR of pEX.RFP, pEX and pMA.His==== | ====Colony PCR of pEX.RFP, pEX and pMA.His==== | ||
Picked four pEX.RFP colonies for PCR verification. Also ran PCR on the pEX and pMA.His clones from 19/8. PCR and gel run by Mimmi (see above). | Picked four pEX.RFP colonies for PCR verification. Also ran PCR on the pEX and pMA.His clones from 19/8. PCR and gel run by Mimmi (see above). | ||
| + | |||
=====Results===== | =====Results===== | ||
*pMA.His clone verified for correct insert. | *pMA.His clone verified for correct insert. | ||
*pEX and pEX.RFP need to be re-run, as they did not result in any bands. | *pEX and pEX.RFP need to be re-run, as they did not result in any bands. | ||
| - | ====Plasmid prep | + | ====ON cultures==== |
| + | *pMA.His | ||
| + | *pEX (not yet verified) | ||
| + | *pEX.RFP 3 (not yet verified) | ||
| + | |||
| + | 3 ml LB + 100 Amp. 30 °C ON. | ||
| + | |||
| + | ===Plasmid prep=== | ||
''From 19/8 ON cultures'' | ''From 19/8 ON cultures'' | ||
| Line 165: | Line 172: | ||
|} | |} | ||
| - | + | ===Assembly of His⋅SOD/yCCS into pSB1K3=== | |
''Step I of C-CPP cloning strategy (19/8)'' | ''Step I of C-CPP cloning strategy (19/8)'' | ||
| - | + | ====Digestions==== | |
| - | [pSB1C3.m-yCCS 1] = 101.2 ng/μl | + | [pSB1C3.m-yCCS 1] = 101.2 ng/μl<br /> |
[pSB1C3.m-SOD 2] = 105.5 ng/μl | [pSB1C3.m-SOD 2] = 105.5 ng/μl | ||
| Line 221: | Line 228: | ||
† ''Miscalculation'' | † ''Miscalculation'' | ||
| - | + | ====Ligations==== | |
Without prior enzyme inactivation or DNA purification. | Without prior enzyme inactivation or DNA purification. | ||
{|border="1" cellpadding="1" cellspacing="0" | {|border="1" cellpadding="1" cellspacing="0" | ||
| - | |align="center"|[μl] | + | |align="center" valign="middle"|[μl] |
!Lig pSB1K3.<br />His⋅SOD | !Lig pSB1K3.<br />His⋅SOD | ||
!Lig pSB1K3.<br />His⋅yCCS | !Lig pSB1K3.<br />His⋅yCCS | ||
| - | |rowspan="9" align="center"|Vector/insert ratio:<br />1:3.<br /><br /> Inc.: 22 °C, 0:10 | + | |rowspan="9" align="center" width="100"|Vector/insert ratio:<br />1:3.<br /><br /> Inc.: 22 °C, 0:10 |
|- | |- | ||
|'''100 ng vector''' | |'''100 ng vector''' | ||
| Line 265: | Line 272: | ||
Enzyme inactivation (not PstI): 80 °C, 20 min. | Enzyme inactivation (not PstI): 80 °C, 20 min. | ||
| - | + | ====Transformation==== | |
Standard transformation protocol. | Standard transformation protocol. | ||
*1 μl ligation mix | *1 μl ligation mix | ||
| Line 272: | Line 279: | ||
*Cells plated onto 50 Km LB agar plates | *Cells plated onto 50 Km LB agar plates | ||
| - | ==== | + | ---- |
| - | + | ==Nina== | |
| - | + | ||
| - | + | ||
| - | 3 ml LB | + | ===Colony PCR on fusion protein=== |
| + | |||
| + | I performed a colony PCR on the dish with fusion proteins. | ||
| + | |||
| + | PCR mix for 5 tubes: | ||
| + | |||
| + | *MgCl 5 ul | ||
| + | *Buffer 5X 50 ul | ||
| + | *dNTP 5 ul | ||
| + | *primer revers for IgG protease 15 ul | ||
| + | *primer VF2 15 ul | ||
| + | *polymerase PjuX7 5 ul | ||
| + | *H2O 150 ul | ||
| + | |||
| + | PCR prgm: | ||
| + | |||
| + | ===Agarose gel on fusion protein=== | ||
| + | |||
| + | I ran the PCR product on an 1 % agarose gel 100 V in order to verify that I have bands corresponding to a correct size of the fusion protein. | ||
| + | |||
| + | Ladder: GeneRuler™ DNA Ladder Mix, ready-to-use, 100-10,000 bp Fermentas | ||
| + | |||
| + | Arrangement on gel: | ||
| + | |||
| + | [[Image:Bild11.jpg]] | ||
| + | |||
| + | [[Image:Bild13.jpg|250px]] | ||
| + | |||
| + | Lane nr 3 looks like it could represent the wanted band of the fusion protein by protein A and IgG protease, which could be about 1300 nt. | ||
| + | |||
| + | |||
| + | ===Overnight culture of fusion protein=== | ||
| + | |||
| + | I inoculated colony nr 3 of the dish with the fusion protein into 12 ml LB and 24 ul chloramphenicol (50 mg/ml). This was incubated in 37 °C in shake overnight. | ||
| + | |||
| + | ===Digestion of protein A=== | ||
| + | |||
| + | I wanted to insert protein A in the peX vector in order to study its overexpression in BL21. I used the sample of protein A that had previously been digested with the restriction enzyme AgeI. | ||
| + | |||
| + | Digestion: | ||
| + | |||
| + | *H2O 15 ul | ||
| + | *10 X fastdigest buffer 2 ul | ||
| + | *DNA 2 ul | ||
| + | *Restriction enzyme XbaI (fastdigest) 1 ul | ||
| + | |||
| + | I incubated in 37 °C for 5 min and then inactivated the restriction enzyme XbaI in 65 °C for 20 min. | ||
| + | |||
| + | ===Digestion of peX vector=== | ||
| + | |||
| + | I digested previously mini prepped peX vector in order for it to be able to ligate with the protein A gene that has been cut with the restriction enzymes AgeI and XbaI. | ||
| + | |||
| + | Digestion: | ||
| + | |||
| + | *H2O 14 ul | ||
| + | *10 X fastdigest buffer 2 ul | ||
| + | *DNA 2 ul | ||
| + | *Restriction enzyme XbaI (fastdigest) 1 ul | ||
| + | *Restriction enzyme AgeI (fastdigest) 1 ul | ||
| + | |||
| + | I incubated in 37 °C for 5 min and then inactivated the restriction enzyme XbaI in 65 °C for 20 min. | ||
| + | |||
| + | ===Agarose gel on digest products=== | ||
| + | |||
| + | I ran the digested products protein A and peX vector on an agarose gel 1 % 100 V in order to perform a gel clean up if the results would be good. This step would give me samples without any interupting bufferts from the digesion when perfoming a ligation in a later stage. | ||
| + | |||
| + | Ladder: GeneRuler™ DNA Ladder Mix, ready-to-use, 100-10,000 bp Fermentas | ||
| + | |||
| + | Arragement on gel: | ||
| + | |||
| + | [[Image:Bild14.jpg]] | ||
| + | |||
| + | [[Image:Bild15.jpg|250px]] | ||
| + | |||
| + | Unfortunately I did not obtain satisfactory results on the digested protein A since I did not see any band representing the cut gene (180 nt)on the gel. In addition I believe the band representing the digested peX is unclear in its meaning. This is because I do not observe any small band that have been cut out of the digested vector. However, the band of the digested peX vector appears to correspond to the correct size after a digestion with AgeI and XbaI. It might just be that the cut site that should appear on the gel is in such a low concentration that it is not visiable on the gel, meaning still that the vector has become digested properly. | ||
| + | |||
| + | I continued with a gel clean up of only the digested peX vector and I re-did a digestion of the gene protein A. The gel clean up of peX started by cutting out the band and leaving it in the refrigerator until I had a proper band of the protein A to cut out too. Then I would perform a complete gel clean up of the two samples peX and protein A. | ||
| + | |||
| + | ===Digestion of protein A=== | ||
| + | |||
| + | I re-do a digestion of protein A, however this time I digest two different samples containing proten A. | ||
| + | |||
| + | Digestion of protein A (18/8): | ||
| + | |||
| + | *H2O 15 ul | ||
| + | *10X fastdigest buffer 2 ul | ||
| + | *DNA 2 ul | ||
| + | *Restriction enzyme XbaI 1 ul | ||
| + | |||
| + | Digestion of protein A (12/8): | ||
| + | |||
| + | *H2O 14 ul | ||
| + | *10X fastdigest buffer 2 ul | ||
| + | *DNA 2 ul | ||
| + | *Restriction enzyme XbaI 1 ul | ||
| + | *Restriction enzyme AgeI 1 ul | ||
| + | |||
| + | Both samples were incubated in 37 °C for 15 min. | ||
| + | |||
| + | ===Agarose gel digest products=== | ||
| + | |||
| + | I ran the two digested protein A samples on an agarose gel 1 % 120 V in order to verify that the gene has been cut and can therefore become ligated with peX. | ||
| + | |||
| + | Ladder: GeneRuler™ DNA Ladder Mix, ready-to-use, 100-10,000 bp Fermentas | ||
| + | |||
| + | Arragement on gel: | ||
| + | |||
| + | [[Image:Bild16.jpg]] | ||
| + | |||
| + | [[Image:Bild17.jpg|250px]] | ||
| + | |||
| + | The gel showed that the digestion of protein A from the sample 12/8 had become cut and therefore I cut out this band for a gel clean up. | ||
| + | |||
| + | ===Gel clean up on peX and protein A=== | ||
| + | |||
| + | I performed a gel clean up on the digested peX vector and the protein A gene. | ||
| + | |||
| + | The procedure was according to the method described in protocols. | ||
| + | |||
| + | *Step 2: peX and protein A were weighed to 200 mg, thus I added 200 mg * 3 = 600 ul QX1 to each sample. | ||
| + | *Step 3: I added 30 ul of QIAEX II to the peX sample and 10 ul to the protein A sample. | ||
| + | *Step 9: i incubated the samples in RT for 5 min. | ||
| + | |||
| + | Spectrophotomer: | ||
| + | |||
| + | [[Image:Bild18.jpg]] | ||
| + | |||
| + | {{Stockholm/Footer}} | ||
Latest revision as of 10:58, 26 October 2010
Contents |
Hassan
midnight update, after a fast recovery from sickness!
Version 0.5.1
Version 0.6.1
Mimmi
pMA
Primer dilution
| pMA_VF | 91µl sH2O --> 100µM | 10µl + 90µl sH2O --> 10µM | ||
| pMA_VR | 91µl sH2O --> 100µM | 10µl + 90µl sH2O --> 10µM |
pMA/pEX
verification PCR
| pEX | pMA | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Mix | (µl) | X6 | X3 | primers | conditions | ||||
| sH2O | 22.5 | 135 | 67.5 | pEX_VF | time | °C | |||
| F primer | 1 | 6 | 3 | pEX_VR | 2m | 94 | |||
| R primer | 1 | 6 | 3 | pMA_VF | 30s | 94 | ) | ||
| DNA | 0.5 | 5X0.5 | 2X0.5 | pMA_VR | 30s | 60 | > 30 cycles | ||
| tot | 25µl | 250µl | 75µl | 2m30s | 72 | ) | |||
| 10m | 72 | ||||||||
| oo | 10 | ||||||||
Andreas
Transformation results
From 19/8 transformations
- pEX.RFP: Good yield of red (positive) and white (negative) colonies.
- pSB1K3.SOD⋅His: Very few colonies
- pSB1K3.yCCS.His: Very few colonies
- Km probably not feasible with quick transformation.
Colony PCR of pEX.RFP, pEX and pMA.His
Picked four pEX.RFP colonies for PCR verification. Also ran PCR on the pEX and pMA.His clones from 19/8. PCR and gel run by Mimmi (see above).
Results
- pMA.His clone verified for correct insert.
- pEX and pEX.RFP need to be re-run, as they did not result in any bands.
ON cultures
- pMA.His
- pEX (not yet verified)
- pEX.RFP 3 (not yet verified)
3 ml LB + 100 Amp. 30 °C ON.
Plasmid prep
From 19/8 ON cultures
- E.Z.N.A. Plasmid Mini Prep kit.
- 70 μl elution volume
| DNA concentrations | ||
|---|---|---|
| Sample | Conc. [ng/μl] | A260/A280 |
| pSB1K3.BBa_J04450 | 103.8 | 1.94 |
| pEX | 55.52 | 1.89 |
| pMA.His | 85.67 | 1.87 |
Assembly of His⋅SOD/yCCS into pSB1K3
Step I of C-CPP cloning strategy (19/8)
Digestions
[pSB1C3.m-yCCS 1] = 101.2 ng/μl
[pSB1C3.m-SOD 2] = 105.5 ng/μl
| [μl] | pMA.His | pSB1C3.m-yCCS 1 | pSB1C3.m-SOD 1 | Inc.: 37 °C, 0:30 (FD) or 1:30 (NgoMIV) |
|---|---|---|---|---|
| 10X FD buffer | 3 | 3 | 3 | |
| dH2O | 6.7† | 6 | 7 | |
| 2 μg DNA | 23.3 | 20 | 19 | |
| FD EcoRI | 0.5 | – | – | |
| FD PstI | – | 1 | 1 | |
| FD AgeI | 0.5 | – | – | |
| NgoMIV | – | 1 | 1 | |
| 30† | 30 | 30 |
† Miscalculation
Ligations
Without prior enzyme inactivation or DNA purification.
| [μl] | Lig pSB1K3. His⋅SOD | Lig pSB1K3. His⋅yCCS | Vector/insert ratio: 1:3. Inc.: 22 °C, 0:10 |
|---|---|---|---|
| 100 ng vector | 2.2 | 2.2 | |
| His insert | 4.0 | 4.0 | |
| SOD insert | 5.1 | – | |
| yCCS | – | 5.7 | |
| 5X Rapid Lig. buf. | 4 | 4 | |
| dH2O | 2.7 | 2.1 | |
| T4 DNA ligase | 1 | 1 | |
| 20 | 20 |
Enzyme inactivation (not PstI): 80 °C, 20 min.
Transformation
Standard transformation protocol.
- 1 μl ligation mix
- Lig pSB1K3.His⋅SOD
- Lig pSB1K3.His⋅yCCS
- Cells plated onto 50 Km LB agar plates
Nina
Colony PCR on fusion protein
I performed a colony PCR on the dish with fusion proteins.
PCR mix for 5 tubes:
- MgCl 5 ul
- Buffer 5X 50 ul
- dNTP 5 ul
- primer revers for IgG protease 15 ul
- primer VF2 15 ul
- polymerase PjuX7 5 ul
- H2O 150 ul
PCR prgm:
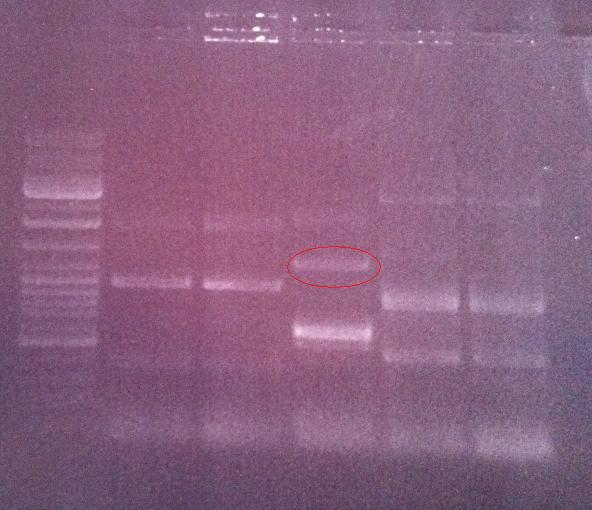
Agarose gel on fusion protein
I ran the PCR product on an 1 % agarose gel 100 V in order to verify that I have bands corresponding to a correct size of the fusion protein.
Ladder: GeneRuler™ DNA Ladder Mix, ready-to-use, 100-10,000 bp Fermentas
Arrangement on gel:
Lane nr 3 looks like it could represent the wanted band of the fusion protein by protein A and IgG protease, which could be about 1300 nt.
Overnight culture of fusion protein
I inoculated colony nr 3 of the dish with the fusion protein into 12 ml LB and 24 ul chloramphenicol (50 mg/ml). This was incubated in 37 °C in shake overnight.
Digestion of protein A
I wanted to insert protein A in the peX vector in order to study its overexpression in BL21. I used the sample of protein A that had previously been digested with the restriction enzyme AgeI.
Digestion:
- H2O 15 ul
- 10 X fastdigest buffer 2 ul
- DNA 2 ul
- Restriction enzyme XbaI (fastdigest) 1 ul
I incubated in 37 °C for 5 min and then inactivated the restriction enzyme XbaI in 65 °C for 20 min.
Digestion of peX vector
I digested previously mini prepped peX vector in order for it to be able to ligate with the protein A gene that has been cut with the restriction enzymes AgeI and XbaI.
Digestion:
- H2O 14 ul
- 10 X fastdigest buffer 2 ul
- DNA 2 ul
- Restriction enzyme XbaI (fastdigest) 1 ul
- Restriction enzyme AgeI (fastdigest) 1 ul
I incubated in 37 °C for 5 min and then inactivated the restriction enzyme XbaI in 65 °C for 20 min.
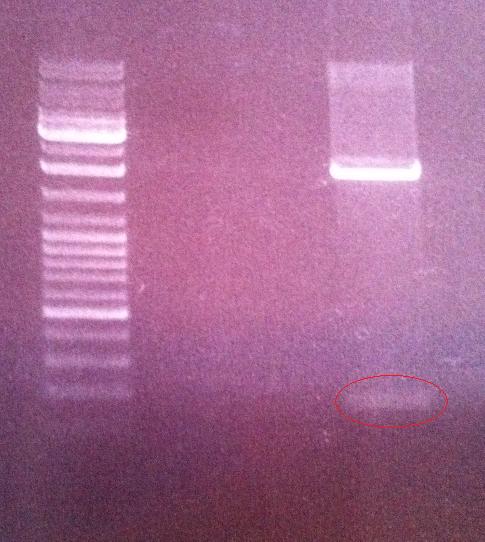
Agarose gel on digest products
I ran the digested products protein A and peX vector on an agarose gel 1 % 100 V in order to perform a gel clean up if the results would be good. This step would give me samples without any interupting bufferts from the digesion when perfoming a ligation in a later stage.
Ladder: GeneRuler™ DNA Ladder Mix, ready-to-use, 100-10,000 bp Fermentas
Arragement on gel:
Unfortunately I did not obtain satisfactory results on the digested protein A since I did not see any band representing the cut gene (180 nt)on the gel. In addition I believe the band representing the digested peX is unclear in its meaning. This is because I do not observe any small band that have been cut out of the digested vector. However, the band of the digested peX vector appears to correspond to the correct size after a digestion with AgeI and XbaI. It might just be that the cut site that should appear on the gel is in such a low concentration that it is not visiable on the gel, meaning still that the vector has become digested properly.
I continued with a gel clean up of only the digested peX vector and I re-did a digestion of the gene protein A. The gel clean up of peX started by cutting out the band and leaving it in the refrigerator until I had a proper band of the protein A to cut out too. Then I would perform a complete gel clean up of the two samples peX and protein A.
Digestion of protein A
I re-do a digestion of protein A, however this time I digest two different samples containing proten A.
Digestion of protein A (18/8):
- H2O 15 ul
- 10X fastdigest buffer 2 ul
- DNA 2 ul
- Restriction enzyme XbaI 1 ul
Digestion of protein A (12/8):
- H2O 14 ul
- 10X fastdigest buffer 2 ul
- DNA 2 ul
- Restriction enzyme XbaI 1 ul
- Restriction enzyme AgeI 1 ul
Both samples were incubated in 37 °C for 15 min.
Agarose gel digest products
I ran the two digested protein A samples on an agarose gel 1 % 120 V in order to verify that the gene has been cut and can therefore become ligated with peX.
Ladder: GeneRuler™ DNA Ladder Mix, ready-to-use, 100-10,000 bp Fermentas
Arragement on gel:
The gel showed that the digestion of protein A from the sample 12/8 had become cut and therefore I cut out this band for a gel clean up.
Gel clean up on peX and protein A
I performed a gel clean up on the digested peX vector and the protein A gene.
The procedure was according to the method described in protocols.
- Step 2: peX and protein A were weighed to 200 mg, thus I added 200 mg * 3 = 600 ul QX1 to each sample.
- Step 3: I added 30 ul of QIAEX II to the peX sample and 10 ul to the protein A sample.
- Step 9: i incubated the samples in RT for 5 min.
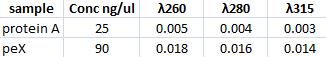
Spectrophotomer:
 |

|
 |

|
 |

|

|

|
 "
"