|
|
| (17 intermediate revisions not shown) |
| Line 2: |
Line 2: |
| | {{:Team:LMU-Munich/Templates/Page Header}} | | {{:Team:LMU-Munich/Templates/Page Header}} |
| | | | |
| - | == Apo-Control ==
| + | [[image:Apologo.png|600px|center|ApoControl logo]] |
| | | | |
| - | The ApoControl team consists of 12 biology students from the undergraduate school of biology at LMU. We are currently undertaking three subprojects on controllable cell-death. The goal is to develop a system to improve the efficiency and specificity of gene expression in eukaryotic cell-lines and more specifically, to select cells expressing the target gene against cells that do not. Here proapoptotic gens instead of antibiotic resistance are used as selection marker to induce clean cell-death at different stimuli. The project is divided into three parts, each representing different approaches to achieve this goal.
| + | == ApoControl == |
| | | | |
| - | ===Cut'N'survive System===
| + | The ApoControl team consists of 12 biology students from the undergraduate school of biology at LMU. We are currently undertaking three subprojects on controllable cell-death. The goal is to develop a system to improve the efficiency and specificity of gene expression in eukaryotic cell-lines and more specifically, to select cells expressing the target gene against cells that do not. Here proapoptotic genes instead of antibiotic resistance are used as selection markers to induce clean cell-death at different stimuli. The project is divided into three parts, each representing different approaches to achieve this goal. |
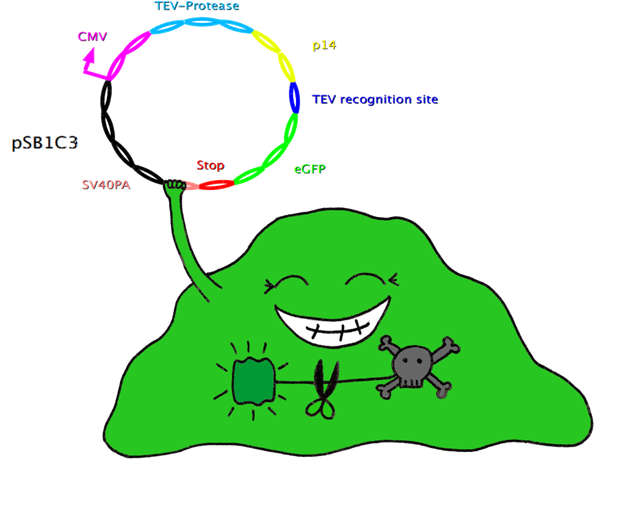
| - | In this system, the apoptosis triggering protein Bak is controlled by the tet-on system for inducible apoptosis. Bak can be degraded when the target gene is successfully expressed, therefore ensuring the survival of the wished cells. Bak is combined with a certain interacting protein p17*, TEV-protease recognition site and N-Degron. This construct should be stably integrated into the cell line genome. The target plasmid consists of eGFP as target gene, another TEV-protease recognition site, the interaction partner of p17* and TEV-protease. When the target plasmid is expressed, the interaction proteins bring the TEV-protease together with its recognition site; TEV cuts Bak, together with N-Degron, free from the rest of the protein complex, where N-Degron with its free N-terminal acts as degradation signal of protease in the cells. Thus this system enables selection of cells expressing the target gene while the cells not transfected undergo induced apoptosis.
| + | |
| | | | |
| - | ===Jump-or-Die System===
| + | [[Image:cnsZelle.png|120px|Logo|right]] |
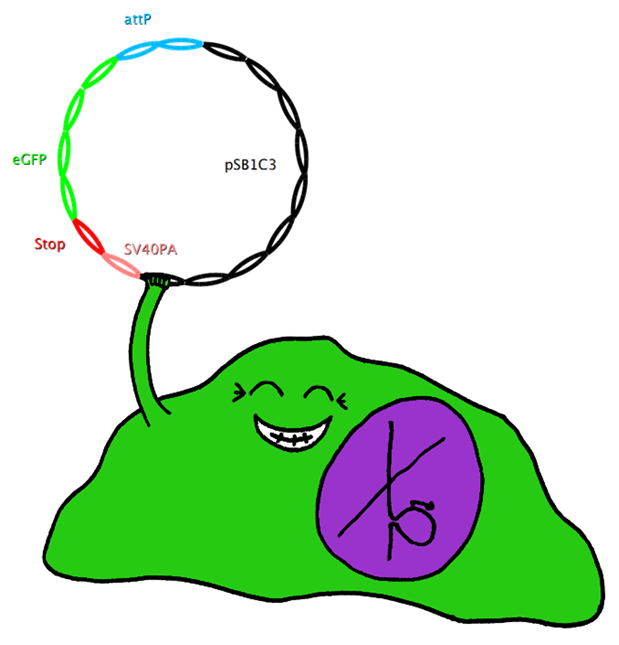
| - | This system also uses Bak as apoptosis inducing selection marker together with the tet-on promoter, which is to be stably integrated in the cell-line genome. However, this construct also includes an upstream bacterial attachment site (attB) and a downstream SV40 polyadenylation site (SV40PA). The target gene eGFP is combined with phage attachment site (attP) and another SV40PA but without promoter. The target plasmid is co-transfected with integrase PhiC31o into the cell line. Therefore the target gene can only be expressed if integrated into the cells’ genome, and the stop-codon and SV40PA following the target gene stop the expression of bak, ensuring the survival of the cell while other cells not expressing the target gene undergo induced apoptosis.
| + | |
| | | | |
| - | ===The ProSearch System=== | + | ===Cut'N'Survive System=== |
| - | This system is to search for new enhancer-promoter combinations that would efficiently trigger gene expression in response to different stimuli. This part of the project is carried out through bioinformatic modeling process. The end-product will be a databank of related genes and translation factors at the input of wished environmental conditions (hypoxia, hyperthermia etc) and the used cell-line. This system will be useful for the construction of new inducible systems trying to utilize apoptosis or other gene expression in human cell-lines.
| + | In this system, the apoptosis triggering protein Bak is controlled by the tet-on system for inducible apoptosis. Bak can be degraded when the target gene is successfully expressed, therefore ensuring the survival of the wished cells. Bak is combined with a certain interacting protein p14*, TEV-protease recognition site and N-Degron. This construct should be stably integrated into the cell line genome. The target plasmid consists of eGFP as a target gene, another TEV-protease recognition site, the interaction partner of p14* (SF3B) and the TEV-protease. When the target plasmid is expressed, the interaction proteins bring the TEV-protease together with its recognition site; TEV cuts Bak, together with N-Degron, free from the rest of the protein complex, where N-Degron with its free N-terminal acts as a degradation signal of protease in the cells. Thus this system enables selection of cells expressing the target gene while the not transfected cells undergo induced apoptosis. |
| | | | |
| - | == Meilensteine ==
| + | [[Image:Jumplogo.png|120px|Logo|right]] |
| | | | |
| - | Für die Projekte haben wir uns jeweils folgende „Meilensteine“ gesetzt:
| + | ===Jump-or-Die System=== |
| - | | + | This system also uses Bak as apoptosis inducing selection marker together with the tet-on promoter, which is to be stably integrated in the cell-line genome. However, this construct also includes an upstream bacterial attachment site (attB) and a downstream SV40 polyadenylation site (SV40PA). The target gene eGFP is combined with phage attachment site (attP) and another SV40PA but without promoter. The target plasmid is co-transfected with integrase PhiC31o into the cell line. Therefore the target gene can only be expressed if integrated into the cells’ genome, and the stop-codon and SV40PA following the target gene stop the expression of bak, ensuring the survival of the cell while other cells not expressing the target gene undergo induced apoptosis. |
| - | === „TEV-System“ === | + | |
| - | | + | |
| - | 1. Verknüpfen von Bak mit interagierendem Protein A, einem N-Degron und einer Schnittstelle für die TEV-Protesase, sodass Bak noch aktiv bleibt. Davor ist ein induzierbarer Promotor und eine Antibiotikaresistenz geschaltet (Nachweis: induzierbarer Zelltod)
| + | |
| - | Aufwand: ca. 6-8 Wochen, 2000€
| + | |
| - | | + | |
| - | 2. Verknüpfen von eGFP als Zielgen mit einer TEV-Schnittstelle, einer TEV-Protease, und interagierendem Protein B. (Nachweis: eGFP leuchtet, Proteinchromatographie)
| + | |
| - | | + | |
| - | Aufwand: ca. 6-8 Wochen, 2000€, Parallele Arbeit ist möglich und notwendig
| + | |
| - | | + | |
| - | 3. Einbringen beider Konstrukte in Zellen (Nachweis: eGFP leuchtet, einige Zellen sterben, Proteinchromatographie)
| + | |
| - | | + | |
| - | Aufwand: ca. 2 Wochen, 1000€
| + | |
| - | | + | |
| - | 4. Parallel wird versucht, das erste Konstrukt stabil ins Zellgenom einzubauen.
| + | |
| - | | + | |
| - | Aufwand: ca. 2 Wochen, 500€
| + | |
| - | | + | |
| - | === „Jump-In-Variety“ ===
| + | |
| - | | + | |
| - | 1. Bau des in die Zelle zu integrierenden Kontrukts: Antibiotikaresistenz mit Promotor, ein weiterer induzierbarer Promotor, Insert-Sequenz 1, proapoptotisches Gen (Bax) (Nachweis: induzierbare Apoptose)
| + | |
| - | | + | |
| - | Aufwand: 3–4 Wochen, 2000€
| + | |
| - | | + | |
| - | 2. Integration des Konstruktes in Zellen (Nachweis: Antibiotikaresistenz nach einigen Tagen)
| + | |
| - | | + | |
| - | Aufwand: 2–4 Wochen, 1500€
| + | |
| - | | + | |
| - | 3. Konstruktion des Zielgenvektors mit Insert-Sequenz 2, eGFP als Zielgen und einem Stopcodon (Nachweis: nicht einzeln möglich)
| + | |
| | | | |
| - | Aufwand: 2 Wochen, 500€<br>
| + | [[Image:prosearchZelle.jpg|120px|Logo|right]] |
| - | Parallele Arbeit ist möglich und notwendig
| + | |
| | | | |
| - | 4. Transfektion der Zellen mit integriertem Konstrukt mit dem Zielgenvektor. Einbau des Zielgens ins Genom (Nachweis: Induzierter Zelltod bei nicht-Integration, Zielgennahweis bei Integration)
| + | ===ProSearch System=== |
| | + | This system is to search for new enhancer-promoter combinations that efficiently trigger gene expression in response to different stimuli. This part of the project is carried out through a bioinformatic modeling process. The end-product is a databank of active genes, their promoters and transcription factors at the input of a wished environmental condition. This system is useful for the construction of new inducible systems trying to utilize apoptosis or other gene expression in the human cell-line 293T. |
| | | | |
| - | Aufwand: 2 Wochen, 1000€
| |
| | | | |
| | | | |


![]()
![]()







![]()
 "
"