Team:Stockholm/13 August 2010
From 2010.igem.org
(→Andreas) |
|||
| (One intermediate revision not shown) | |||
| Line 1: | Line 1: | ||
{{Stockholm/Top2}} | {{Stockholm/Top2}} | ||
| + | |||
| + | __TOC__ | ||
| + | |||
==Nina== | ==Nina== | ||
| Line 100: | Line 103: | ||
====Gel verification==== | ====Gel verification==== | ||
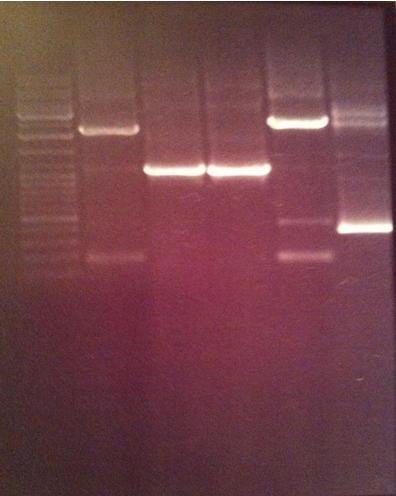
| - | [[image:ColPCR_SOD-yCCS_13aug.png|200px|thumb|Colony PCR verification of mutagenized yCCS and SOD.<br>1 % agarose, 110 V, 40 min.<br>''' | + | [[image:ColPCR_SOD-yCCS_13aug.png|200px|thumb|'''Colony PCR verification of mutagenized yCCS and SOD.'''<br>1 % agarose, 110 V, 40 min.<br>'''1st row:'''1 kb λ, SA1, SA2, SA3, SA4, SC1, SC2, SC3, SC4, SC5, d.yC5<br> |
| - | ''' | + | '''2nd row:''' 1 kb λ, yC1, d.yC1, yC2, d.yC2, yC3, d.yC3, yC4, d.yC4, yC5, PC<br>λ: DNA ladder. d.:digested sample.]] |
1 % agarose, 110 V, 40 min | 1 % agarose, 110 V, 40 min | ||
| Line 107: | Line 110: | ||
=====Results===== | =====Results===== | ||
Due to very unclear and weak bands, I decided to discard the gel and run a new on Monday. | Due to very unclear and weak bands, I decided to discard the gel and run a new on Monday. | ||
| + | |||
| + | {{Stockholm/Footer}} | ||
Latest revision as of 10:50, 26 October 2010
Contents |
Nina
Gel verification of protein A (ZZ domain) and IgG protease
I ran a new agarose gel 1 % on my protein A and IgG protease samples. However I didn't load the positive protein A control sample since I realized that it would not fullfill its purpose considering the vector verification primer I used does not bind to the vector protein A sits in, which is the original vector I obtained it in.
Ladder: GeneRuler™ DNA Ladder Mix, ready-to-use, 100-10,000 bp Fermentas
Arrangement on gels:
Colony-PCR of site-directed mutagenesis Tyrosinase
Colony nr: 2, 4, 6 & 8.
PCR Mix:
Total volume 100 ul, aliquate 25 ul in the four pcr tubes.
- F primer 50 uM 1 ul
- R primer 50 uM 1 ul
- dNTP 10 uM 1 ul
- Buffer phusion 5X 20 ul
- Phusion polymerase 1 ul
- H2O 75 ul
PCR prgm:
98 °C 2 min
22 cycles of:
- 98 °C 30 sec
- 50 °C 30 sec
- 72 °C 2 min
72 °C 1 min
4 °C ∞
I ran 5 ul of the PCR products with 1 ul loading dye 6X on an agarose gel 1 % 80 V.
Ladder: GeneRuler™ DNA Ladder Mix, ready-to-use, 100-10,000 bp Fermentas
Arrangement on gels:
Overnight culture of protein A (ZZ domain) and Tyrosinase
- I inoculated protein A ZZ domain inserted in the bank vector C (with chloramphenicol) colony # 5 in 12 ml LB and 24 ul chloramphenicol 50 mg/ml. This was incubated O/N in 37 °C with shake.
- I inoculated tyrosinase in its original vector colonies # 2, 4, 6 & 8 in 12 ml LB and 24 ul ampicillin 50 mg/ml. This was incubated O/N in 37 °C with shake.
Sequencing of protein A (ZZ domain)
I prepared a tube of protein A ZZ domain for sequencing:
15 ul plasmid from a mini-prep and 1.5 ul (10uM) primer. Preferably the forward primer (VF) of the vectors verification primers.
- ASB0045 303
Andreas
Site-directed mutagenesis of SOD & yCCS
Transformation results
- SOD-->Amp 100: Few colonies, probably due to contaminated competent cells.
- SOD-->Cm 25: Good colony yield
- yCCS-->Cm 25: Good colony yield
Colony PCR
- SOD-->Amp: 4 colonies (SA1-SA4)
- SOD-->Cm: 5 colonies (SC1-SC5)
- yCCS-->Cm: 5 colonies (yC1-yC5)
- Positive control: pSB1C3.BBa_J18932 plasmid (PC)
Procedures as described in the colony PCR protocol. Elongation time: 1:40
yCCS digestion
Amplified yCCS DNA was digested to verify successful site-directed mutagenesis (i.e. EcoRI site removal).
- 2 μl 10X FD Green buffer
- 17 μl DNA
- 1 μl FastDigest EcoRI
Incubation: 37 °C, 10 min
Gel verification
1 % agarose, 110 V, 40 min
Results
Due to very unclear and weak bands, I decided to discard the gel and run a new on Monday.
 |

|
 |

|
 |

|

|

|
 "
"