Team:Stockholm/11 August 2010
From 2010.igem.org
NinaSchiller (Talk | contribs) (→Overnight culture of protein A (ZZ domain)) |
|||
| (2 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Stockholm/Top2}} | {{Stockholm/Top2}} | ||
| + | |||
__TOC__ | __TOC__ | ||
| Line 113: | Line 114: | ||
Digested vector and insert will be tested on gel tomorrow. | Digested vector and insert will be tested on gel tomorrow. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ---- | ||
| + | |||
| + | == Mimmi == | ||
| + | |||
| + | === MITF === | ||
| + | ==== pRc/CMV plasmid preparation ==== | ||
| + | |||
| + | <u>Plasmid Mini Kit I, protocol I</u> | ||
| + | |||
| + | *Spinn down cells | ||
| + | **2X5ml | ||
| + | *Eluate in 300µl | ||
| + | **2X175µl | ||
| + | *Transfer to 1.5ml tube | ||
| + | **merge the two fractions | ||
| + | *Wash 2X with wash buffer | ||
| + | *Eluate with 50µl sH<sub>2</sub>O | ||
| + | |||
| + | |||
| + | |||
| + | ==== Controling the pRc/CMV plasmid ==== | ||
| + | |||
| + | ::*Cut with restriction enzymes: | ||
| + | :::#HindIII + SmaI | ||
| + | :::#ApaI + SmaI | ||
| + | :::#uncut plasmid | ||
| + | {|align="center" | ||
| + | ! Mix | ||
| + | ! (µl) X2 tubes | ||
| + | |- | ||
| + | | sH<sub>2</sub>O | ||
| + | | 22 | ||
| + | |- | ||
| + | | 10Xbuffer | ||
| + | | 3 | ||
| + | |- | ||
| + | | DNA | ||
| + | | 3 | ||
| + | |- | ||
| + | | enzyme | ||
| + | | 2X1 | ||
| + | |- | ||
| + | | align="right" | tot | ||
| + | | 30µl | ||
| + | |} | ||
| + | |||
| + | ::*Keep in heatblock, 37°C, 10min | ||
| + | |||
| + | [[Image:2010-08-11_pRcCMV.MITF_restriction_enzymes_1.jpg|200px|thumb|left|]] | ||
| + | '''GEL''' | ||
| + | {| | ||
| + | ! well | ||
| + | ! sample | ||
| + | |- | ||
| + | | 1 | ||
| + | | ladder | ||
| + | |- | ||
| + | | 2 | ||
| + | | HindIII+SmaI | ||
| + | |- | ||
| + | | 3 | ||
| + | | ApaI+SmaI | ||
| + | |- | ||
| + | | 4 | ||
| + | | plasmid control | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ::*Cut with restriction enzymes: | ||
| + | :::#BglII + XbaI | ||
| + | :::#HindIII | ||
| + | :::#XbaI | ||
| + | |||
| + | {| align="center" | ||
| + | ! Mix | ||
| + | ! (µl) X2 tubes | ||
| + | |- | ||
| + | | sH<sub>2</sub>O | ||
| + | | 22 | ||
| + | |- | ||
| + | | 10Xbuffer | ||
| + | | 3 | ||
| + | |- | ||
| + | | DNA | ||
| + | | 3 | ||
| + | |- | ||
| + | | enzyme | ||
| + | | 2X1 | ||
| + | |- | ||
| + | | align="right" | tot | ||
| + | | 30µl | ||
| + | |} | ||
| + | |||
| + | ::*Keep in heatblock, 37°C, 10min | ||
| + | |||
| + | |||
| + | [[Image:2010-08-16_pRcCMV.MITF_restriction_enzymes_1.jpg|200px|thumb|left|]] | ||
| + | '''GEL''' | ||
| + | {| | ||
| + | ! well | ||
| + | ! sample | ||
| + | |- | ||
| + | | 1 | ||
| + | | ladder | ||
| + | |- | ||
| + | | 2 | ||
| + | | BglII + XbaI | ||
| + | |- | ||
| + | | 3 | ||
| + | | HindIII | ||
| + | |- | ||
| + | | 4 | ||
| + | | XbaI | ||
| + | |} | ||
| + | |||
| + | {{Stockholm/Footer}} | ||
Latest revision as of 10:49, 26 October 2010
Contents |
Hassan
- Started making animation for what's happening in our project.
- TODO for tomorrow: complete the interaction map, and starting summarizing texts for the map, hope to finish it by sunday...
Nina
Digestion of Tyrosinase
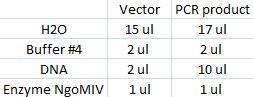
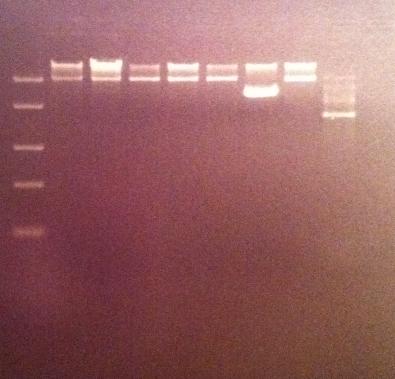
I cut the miniprepped site directed mutagenesis and PCR product tyrosinase with the restriction enzyme NgoMIV.
Digestion:
Incubated in 37 °C for 2 hours.
I ran the digested products in an agarose 1 % gel 80 V.
DNA Ladder: FastRuler™ Middle Range, ready-to-use, 100-5000 bp Fermentas
Arrangement on gel:
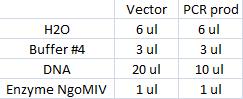
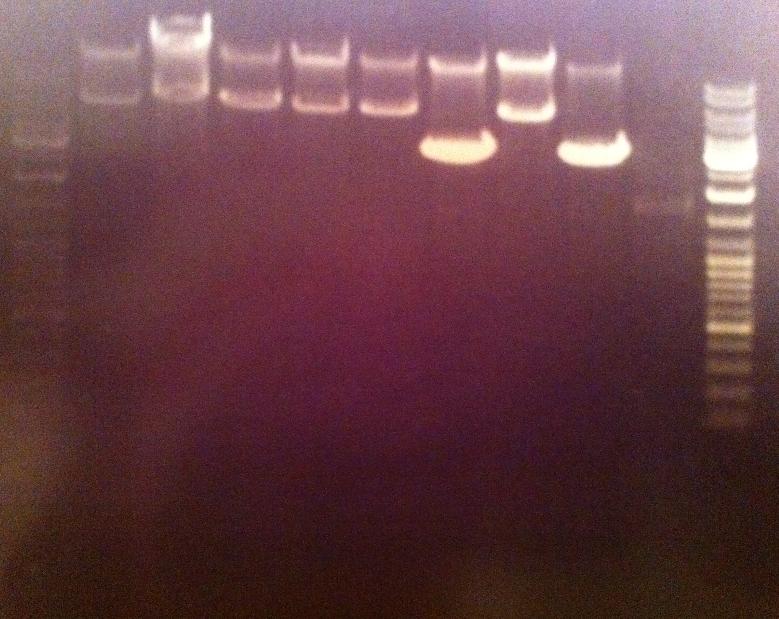
I ran a new digestion since the first one did not indicate wether or not the digest had worked or not. The second digest I altered the amounts in the mixture into those values that I have used previously when digesting.
Second digest:
Incubated in 37 °C for 1 hour (PCR prod) and 3 hours (vector).
I ran the digested products in an agarose 1 % gel 80 V.
Ladder: GeneRuler™ DNA Ladder Mix, ready-to-use, 100-10,000 bp Fermentas
Arrangement on gel:
I still don't get reliable results that the enzyme has cut, therefore I will run a gel tomorrow with an uncut vector to compare with the cut vector sample, this should show if the enzyme can cut and from there I know that the site directed mutagenesis tyrosinase samples have been succesful in altering the restiction site for NgoMIV since they won't be cut.
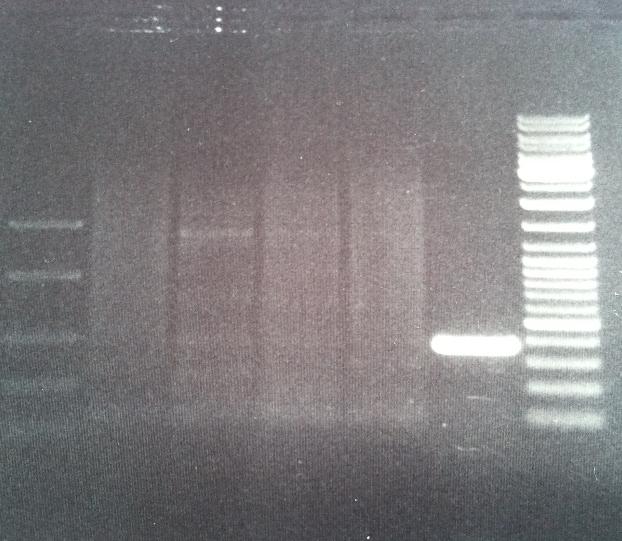
Colony PCR of protein A (ZZ domain)
I ran a colony PCR on the transformed protein A ZZ domain in the bank vector C in order to verify that I have inserted the gene correct in the vector.
PCR master mix (calculated for 7 tubes):
- MgCl2 7 ul
- Buffer phusion 5X 70 ul
- dNTP 10 uM 7ul
- primer VR 10 uM 21 ul
- primer Forward 10 uM 21 ul
- polymerase PjuX7 7 ul
- H2O 210 ul
5 ul of the PCR products with 1 ul loading dye 6X were run on an agarose 1 % gel, 80 V.
Ladder to the right: GeneRuler™ DNA Ladder Mix, ready-to-use, 100-10,000 bp Fermentas
Ladder to the left: GeneRuler™ Low Range DNA Ladder, 25-700 bp Fermentas
Arrangement on gel:
The lane number 5 looks good and verifys that I have protein A ZZ domain in the bank vector with chrloramphenicol, therefore I will continue with this colony (#5) when working with protein A ZZ in the bank vector.
Overnight culture of protein A (ZZ domain)
I inoculated three falcon tubes containing 12 ml LB and 24 ul chloramphenicol (50mg/ml) with the colony number 5 from the protein A ZZ domain dish. This was incubated overnight in 37 °C with shake. These samples will be miniprepped tomorrow.
Andreas
Cloning of IgG protease
Continued from 10/8
Colony PCR
Four clones picked from each of the two plates (Q for quick transformation; S for standard transformation).
- QA, QB, QC, QD
- SA, SB, SC, SD
- Pos. control: pSB1A3.BBa_J04450 (plasmid)
Procedures
As described in colony PCR protocol. Elongation time: 1:40.
Gel verification
1 %, 80 V, 45 min
Expected: 1262 bp (BBa_J04450: 1386 bp)
Results
Very strangely sized bands again! None of the bands corresponding to the 1262 bp expected. Control corresponding well to expected 1386 bp.
Digested vector and insert will be tested on gel tomorrow.
Mimmi
MITF
pRc/CMV plasmid preparation
Plasmid Mini Kit I, protocol I
- Spinn down cells
- 2X5ml
- Eluate in 300µl
- 2X175µl
- Transfer to 1.5ml tube
- merge the two fractions
- Wash 2X with wash buffer
- Eluate with 50µl sH2O
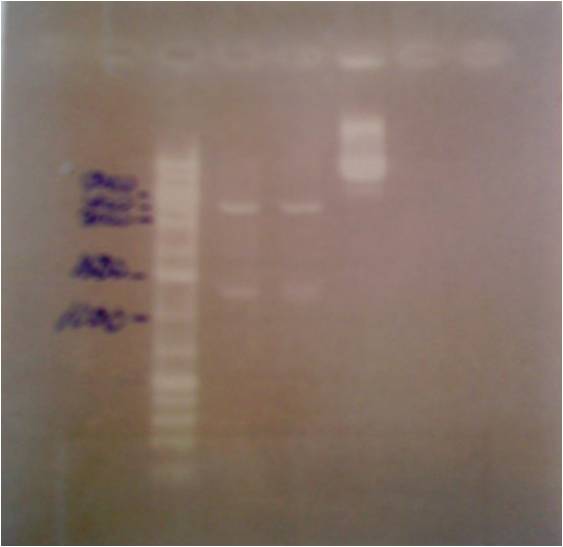
Controling the pRc/CMV plasmid
- Cut with restriction enzymes:
- HindIII + SmaI
- ApaI + SmaI
- uncut plasmid
| Mix | (µl) X2 tubes |
|---|---|
| sH2O | 22 |
| 10Xbuffer | 3 |
| DNA | 3 |
| enzyme | 2X1 |
| tot | 30µl |
- Keep in heatblock, 37°C, 10min
GEL
| well | sample |
|---|---|
| 1 | ladder |
| 2 | HindIII+SmaI |
| 3 | ApaI+SmaI |
| 4 | plasmid control |
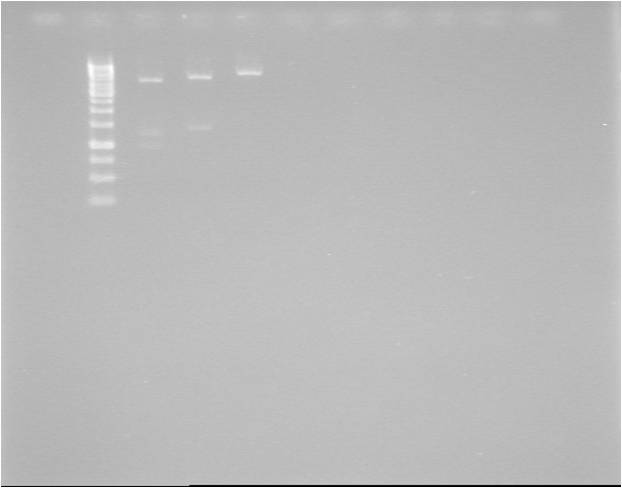
- Cut with restriction enzymes:
- BglII + XbaI
- HindIII
- XbaI
| Mix | (µl) X2 tubes |
|---|---|
| sH2O | 22 |
| 10Xbuffer | 3 |
| DNA | 3 |
| enzyme | 2X1 |
| tot | 30µl |
- Keep in heatblock, 37°C, 10min
GEL
| well | sample |
|---|---|
| 1 | ladder |
| 2 | BglII + XbaI |
| 3 | HindIII |
| 4 | XbaI |
 |

|
 |

|
 |

|

|

|
 "
"