Team:LMU-Munich/TEV-System/Functional Principle
From 2010.igem.org
(Difference between revisions)
(→Selection) |
|||
| (5 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:LMU-Munich/Templates/Page Header}} | {{:Team:LMU-Munich/Templates/Page Header}} | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
== TEVdegron-System == | == TEVdegron-System == | ||
| - | The TEVdegron-System uses and | + | The TEVdegron-System uses and combines several proteins with different properties to select the incorporation of a plasmid by apoptosis. We need two DNA constructs and one of them (construct 1) should be integrated in the cell's DNA. |
=== Construct 1 === | === Construct 1 === | ||
| Line 21: | Line 12: | ||
[[Image:TEV1.jpg|400pxs|TEV Construct 1: Inserted in celline]] | [[Image:TEV1.jpg|400pxs|TEV Construct 1: Inserted in celline]] | ||
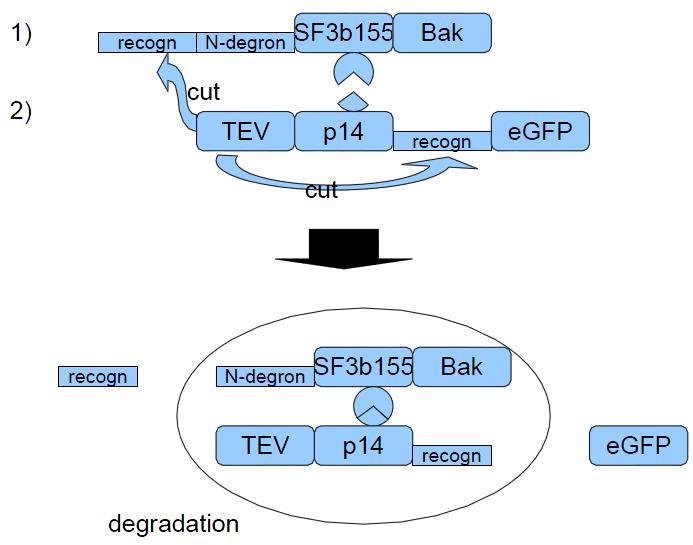
| - | To check the integration of | + | To check the integration of this construct into cellular DNA we need the hygromycin resistance which can be read-off thanks to the CMV-promoter. The TEV-recognition site will be cut by the TEV-protease which is part of construct 2. Due to the scission of TEV-recognition site the N-terminus of N-degron is free which is a signal for the degeneration of this protein. SF3b155 is a protein interacting with p14 from construct 2. This interaction should make sure that the TEV-protease of construct 2 will really "find" the recognition site. The human bak is a apoptosis triggering membrane protein. |
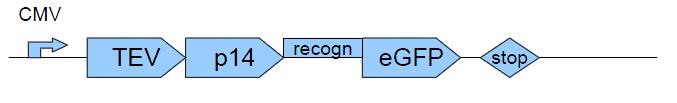
=== Construct 2 === | === Construct 2 === | ||
| Line 29: | Line 20: | ||
[[Image:TEV2.jpg|400pxs|TEV Construct 2: Contains gene of interest]] | [[Image:TEV2.jpg|400pxs|TEV Construct 2: Contains gene of interest]] | ||
| - | The CMV-promoter is to read off construct 2. The task of TEV-protease is to cut TEV-recognition sites, | + | The CMV-promoter is to read off construct 2. The task of TEV-protease is to cut TEV-recognition sites, especially the one ahead of N-degron to signal protein degradation. p14 is interacting with SF3b155 and so increases the rate of finding the TEV-recognition site by the TEV-proteases. eGFP is our example of a gene of interest which can be verified by green flourescence. |
=== Selection === | === Selection === | ||
| - | + | A cellline which has integrated construct 1 and is transfected with construct 2 leads to two outgoings: | |
| + | |||
a) the plasmid has not been incorporated | a) the plasmid has not been incorporated | ||
| Line 39: | Line 31: | ||
b) the plasmid has been incorporated | b) the plasmid has been incorporated | ||
| - | + | These lead to the following consequences. | |
==== Case a) the plasmid has not been incorporated ==== | ==== Case a) the plasmid has not been incorporated ==== | ||
| - | + | After induction of the tet-on promoter construct 1 will be translated into protein. The bak-part will integrate into mitochondrial membrane and as a result will induce apoptosis. | |
==== Case b) the plasmid has been incorporated ==== | ==== Case b) the plasmid has been incorporated ==== | ||
| - | Construct 2 will | + | Construct 2 will be read off and already existing when tet-on promoter will be induced to create construct 1 as protein. Therefore TEV proteases will instantly seperate eGFP from the remain of the protein and will free the N-terminus of N-degron. This will cause the degradation of the whole protein complex - including bak. So the cell will survive and only the gene of interest will be left. |
| + | |||
| + | [[Image:TEV4.jpg|400pxs|TEVdegron-System: TEV-degron-System]] | ||
| + | |||
| + | |||
{{:Team:LMU-Munich/Templates/Page Footer}} | {{:Team:LMU-Munich/Templates/Page Footer}} | ||
Latest revision as of 10:29, 16 August 2010


![]()
![]()







![]()
 "
"