Team:Michigan/Quorum Sensing
From 2010.igem.org
| (24 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
{{Michigan Header}} | {{Michigan Header}} | ||
| - | |||
{|style="color:#1c2bf2;background-color:#fafa19;font-size:9pt;text-align:center" cellpadding="5" cellspacing="0" border="1" bordercolor="#fff" width="62%" | {|style="color:#1c2bf2;background-color:#fafa19;font-size:9pt;text-align:center" cellpadding="5" cellspacing="0" border="1" bordercolor="#fff" width="62%" | ||
|- | |- | ||
| Line 75: | Line 74: | ||
| - | | - | ||
| - | | - | ||
| + | |- | ||
| + | !Week 8 | ||
| + | | - | ||
| + | | - | ||
| + | | - | ||
| + | | - | ||
| + | | [[Team:Michigan/Quorum_Sensing#8/19/2010|8/19/2010]] | ||
| + | | [[Team:Michigan/Quorum_Sensing#8/20/2010|8/20/2010]] | ||
| + | | - | ||
| + | |- | ||
| + | !Week 9 | ||
| + | | [[Team:Michigan/Quorum_Sensing#8/22/2010|8/22/2010]] | ||
| + | | [[Team:Michigan/Quorum_Sensing#8/23/2010|8/23/2010]] | ||
| + | | - | ||
| + | | - | ||
| + | | [[Team:Michigan/Quorum_Sensing#8/26/2010|8/26/2010]] | ||
| + | | [[Team:Michigan/Quorum_Sensing#8/27/2010|8/27/2010]] | ||
| + | | [[Team:Michigan/Quorum_Sensing#8/28/2010|8/28/2010]] | ||
| + | |- | ||
|} | |} | ||
| + | |||
| + | {|cellspacing=0 style="background: transparent" | ||
| + | |-valign="top" | ||
| + | |width="700px" | | ||
| + | |||
=Quorum Sensing Team= | =Quorum Sensing Team= | ||
| Line 138: | Line 161: | ||
**AI-2 reporter & LuxS null mutant (''E. coli'', does not produce AI-2, amp and kan-resistant) | **AI-2 reporter & LuxS null mutant (''E. coli'', does not produce AI-2, amp and kan-resistant) | ||
Removed these strains from 4°C -- they are stored in soft agar stab cultures | Removed these strains from 4°C -- they are stored in soft agar stab cultures | ||
| + | |||
Made one streak plate on LB+amp for each strain from stabs | Made one streak plate on LB+amp for each strain from stabs | ||
| - | *placed in | + | *placed in 37°C (rm. 1239) |
Made one 2mL LB+amp broth cultures in a 15mL tube for each strain from stabs | Made one 2mL LB+amp broth cultures in a 15mL tube for each strain from stabs | ||
*placed in 30°C, 200 rpm shaking (rm. 1230) | *placed in 30°C, 200 rpm shaking (rm. 1230) | ||
| Line 179: | Line 203: | ||
*stored in iGEM box -- -80°C Lin Lab | *stored in iGEM box -- -80°C Lin Lab | ||
Plated LuxS<sup>-</sup> and MarC<sup>-</sup> mutants and ''Pseudomonas'' strains on LB | Plated LuxS<sup>-</sup> and MarC<sup>-</sup> mutants and ''Pseudomonas'' strains on LB | ||
| + | |||
Plated W3110 and MDAI2 on LB+amp+kan | Plated W3110 and MDAI2 on LB+amp+kan | ||
*placed all six plates in 37°C ERB 1239 | *placed all six plates in 37°C ERB 1239 | ||
| Line 368: | Line 393: | ||
**W3110: '''204.86''' | **W3110: '''204.86''' | ||
**MDAI2: '''198.34''' | **MDAI2: '''198.34''' | ||
| - | ''OK, | + | ''OK, I don't know if these have GFP or what. At any rate, if they do, the MDAI2 is too close to W3110. We'll probably have to work with the YFP biobrick instead.'' |
Got ''C. vulgaris'' from Bobby Levine | Got ''C. vulgaris'' from Bobby Levine | ||
| Line 387: | Line 412: | ||
Autoclaved waste flasks | Autoclaved waste flasks | ||
*55 min (30 sterilize) | *55 min (30 sterilize) | ||
| - | * | + | *25°C |
| + | |||
| + | == 8/3/2010 == | ||
| + | ''Alex and Marcus'' | ||
| + | |||
| + | Took yesterday's overnights to Alex's (Xi) lab since the Lin lab fluorospectrophotometer was in use. | ||
| + | *Excitation Frequency: 485 | ||
| + | *Emission Frequency: 545 | ||
| + | *Sensitivity: 50 | ||
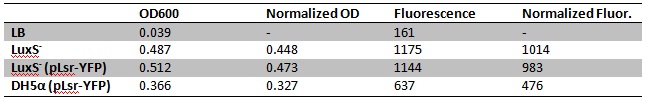
| + | [[Image:LuxS- test.jpg]] | ||
| + | |||
| + | Fluorescence/OD600: | ||
| + | *LuxS<sup>-</sup>: 2263 | ||
| + | *DH5α + pLsr-YFP: 2078 | ||
| + | *LuxS<sup>-</sup> + pLsr-YFP: 1456 | ||
| + | |||
| + | Very confusing results, LuxS<sup>-</sup> was supposed to be the negative control, and shows the highest fluorescence per OD. Some factor must be confounding our assay. | ||
| + | |||
| + | Also, made 10 mL overnights of LuxS<sup>-</sup> (pLsr-YFP) and MDAI2 (pET6 + pET-GFP) in 50 mL tubes at 7 pm in LB+Amp+Kan. Put in shaker at 30°C. | ||
== 8/4/2010 == | == 8/4/2010 == | ||
| Line 495: | Line 538: | ||
<!--Alex, please stop adding your timestamp. Thanks.--> | <!--Alex, please stop adding your timestamp. Thanks.--> | ||
| + | |||
| + | == 8/19/2010 == | ||
| + | ''Alex'' | ||
| + | |||
| + | Acquired new strains (soft agar stabs from Tsao Lab) | ||
| + | *W3110 + pTC6 + pET-GFP | ||
| + | *MDAI2 + pTC6 + pET-GFP | ||
| + | *BL21 + pTC5 + pET-GFP | ||
| + | **produces more AI-2 than W3110 | ||
| + | *moved all to ERB 4°C | ||
| + | Made broth cultures of MDAI2 and BL21 from stab cultures | ||
| + | *each 2 mL LB in a 15mL tube | ||
| + | *37°C, 200 rpm shaking | ||
| + | *placed in incubator at 8pm | ||
| + | |||
| + | Added [[Team:Michigan/Project|Quorum Sensing project description]] to Wiki | ||
| + | |||
| + | == 8/20/2010 == | ||
| + | ''Alex'' | ||
| + | |||
| + | [Earlier, Marcus cryostored BL21 and MDAI2 in Lin -80°C and made a spread plate of new MDAI2 strain.] | ||
| + | |||
| + | Made streak plate of BL21 | ||
| + | *placed in 35°C incubator | ||
| + | Made frozen stock of BL21 and MDAI2 | ||
| + | *each 500 μL culture + 500 μL 50% glycerol | ||
| + | *stored in ERB -20°C | ||
| + | Updated strain database | ||
| + | |||
| + | == 8/22/2010 == | ||
| + | ''Alex'' | ||
| + | |||
| + | Moved MDAI2 spread plate and BL21 streak plate from 37°C to 4°C | ||
| + | |||
| + | ---- | ||
| + | Made BL21 broth culture | ||
| + | *10 mL LB+100μg/mL amp+50μg/mL kan in a 50mL tube | ||
| + | *37°C, 200 rpm shaking | ||
| + | *8:00pm | ||
| + | |||
| + | == 8/23/2010 == | ||
| + | ''Marcus'' | ||
| + | |||
| + | Measured OD600 of overnight in Gulari Lab spectrophotometer. | ||
| + | -Used LB+ Amp(100μg/mL)+ Kan(50μg/mL) blank | ||
| + | -OD was 1.55 | ||
| + | |||
| + | Made dilution to OD 0.02 into 50 mL flask using [[Media:OD600 Cell Dilution Protocol.pdf|OD600 Cell Dilution Protocol]]. | ||
| + | -48.3 mL LB | ||
| + | -1 mL 40% glucose solution | ||
| + | -0.645 mL overnight culture | ||
| + | |||
| + | Put in 37°C shaker for 4 hours and then collected supernatant according to [[Media:QS_Procedures-1-.pdf|Lsr Circuit Test Protocols]]. Supernatant was labeled and stored in the ERB -20°C. | ||
| + | |||
| + | == 8/26/2010 == | ||
| + | ''Alex and Marcus'' | ||
| + | |||
| + | Made cultures of MDAI2 + pTC6 + pET-GFP (new) and LuxS<sup>-</sup> + pLsrA-YFP | ||
| + | *each 2 mL LB + 100μg/mL amp + 50μg/mL kan in each of four 15mL tubes (8 tubes total) | ||
| + | *incubated 30°C, 200 rpm shaking, 9:15pm | ||
| + | |||
| + | == 8/27/2010 == | ||
| + | ''Alex'' | ||
| + | |||
| + | Obtained cultures and supernatants from ERB -- 1:15pm (16 hrs); transfered to Xi Lab, SPH | ||
| + | |||
| + | Began Quorum Sensing experiment again, following [[Media:QS_Procedures-1-.pdf|posted protocol]] | ||
| + | *Used BL21 supernatant instead of MDAI2 | ||
| + | *had to wait until 6:45pm to run exp; cells were overgrown and started to die/pellet | ||
| + | **therefore, took only broth, avoiding pellet, for OD and spin | ||
| + | *spun at 5000rpm instead of 4200 | ||
| + | *plate template: | ||
| + | {| | ||
| + | !well | ||
| + | !pellet | ||
| + | !supernatant | ||
| + | |- | ||
| + | |A1 | ||
| + | |MDAI2 | ||
| + | |LB | ||
| + | |- | ||
| + | |A2 | ||
| + | |MDAI2 | ||
| + | |W3110 | ||
| + | |- | ||
| + | |A3 | ||
| + | |MDAI2 | ||
| + | |BL21 | ||
| + | |- | ||
| + | |A4 | ||
| + | |MDAI2 | ||
| + | |''C. vulgaris'' | ||
| + | |- | ||
| + | |B1 | ||
| + | |LuxS<sup>-</sup>+pLsrA-YFP | ||
| + | |LB | ||
| + | |- | ||
| + | |B2 | ||
| + | |LuxS<sup>-</sup>+pLsrA-YFP | ||
| + | |W3110 | ||
| + | |- | ||
| + | |B3 | ||
| + | |LuxS<sup>-</sup>+pLsrA-YFP | ||
| + | |BL21 | ||
| + | |- | ||
| + | |B4 | ||
| + | |LuxS<sup>-</sup>+pLsrA-YFP | ||
| + | |''C. vulgaris'' | ||
| + | |- | ||
| + | |C1 | ||
| + | |[blank] | ||
| + | |LB | ||
| + | |- | ||
| + | |C2 | ||
| + | |[blank] | ||
| + | |MDAI2 | ||
| + | |- | ||
| + | |C3 | ||
| + | |[blank] | ||
| + | |W3110 | ||
| + | |- | ||
| + | |C4 | ||
| + | |[blank] | ||
| + | |''C. vulgaris'' | ||
| + | |} | ||
| + | |||
| + | *ran growth curve for 16 hrs (overnight) instead of 6, drawing from Singapore protocol | ||
| + | Returned supernatants to ERB -20°C | ||
| + | |||
| + | == 8/28/2010 == | ||
| + | ''Alex'' | ||
| + | |||
| + | Obtained data from QS main experiment | ||
| + | *uploaded [[media:QS_exp_2.xls|here]] | ||
| + | |||
| + | Only the wells with LB added to pellet had a significant increase in fluorescence over time. | ||
| + | *This doesn't make sense, but seems to replicate earlier results. | ||
| + | It seems that the LB blank well was contaminated slightly. This shouldn't really matter, unless the contamination was also in the other two LB wells, in which case maybe co-culture was what cause the increase in fluorescence. However, OD also only increased significantly for the LB wells, so this is probably why the fluorescence also increased. | ||
| + | This does present another experiment idea, however: maybe we can try to culture MDAI2 alone, and then in co-culture with K12 to see if there it a difference in GFP. | ||
| + | At any rate, I'm not sure it's feasible to co-culture with algae, so quorum sensing project is probably done. | ||
| + | |||
| + | |width="250px" style="background: transparent"| | ||
| + | |||
| + | ==='''In the Lab'''=== | ||
| + | |||
| + | [[Image:QS01.jpg|middle|250px]] | ||
| + | |||
| + | [[Image:QS02.jpg|middle|250px]] | ||
| + | |||
| + | [[Image:QS03.jpg|middle|250px]] | ||
| + | |||
| + | [[Image:QS04.jpg|middle|250px]] | ||
| + | |||
| + | [[Image:QS05.jpg|middle|250px]] | ||
| + | |||
| + | [[Image:QS06.jpg|middle|250px]] | ||
| + | |||
| + | [[Image:QS07.jpg|middle|250px]] | ||
Latest revision as of 01:14, 27 October 2010
| Sunday | Monday | Tuesday | Wednesday | Thursday | Friday | Saturday | |
| Week 1 | - | - | - | - | - | - | - |
| Week 2 | - | - | - | 7/7/2010 | 7/8/2010 | - | - |
| Week 3 | - | - | - | - | - | - | - |
| Week 4 | 7/17/2010 | 7/19/2010 | - | 7/21/2010 | 7/22/2010 | 7/23/2010 | - |
| Week 5 | 7/25/2010 | 7/26/2010 | 7/27/2010 | 7/28/2010 | - | 7/30/2010 | - |
| Week 6 | - | 8/2/2010 | - | 8/4/2010 | 8/5/2010 | - | - |
| Week 7 | - | - | - | - | - | - | - |
| Week 8 | - | - | - | - | 8/19/2010 | 8/20/2010 | - |
| Week 9 | 8/22/2010 | 8/23/2010 | - | - | 8/26/2010 | 8/27/2010 | 8/28/2010 |
|
Quorum Sensing TeamMembers include Alex Pyden, Marcus Lehr, Jennifer Hong, Eric Raynal, Katie Miskovich, and Audra Williams 7/7/2010Jennifer and Alex - in Lin Lab Made CaCl2 stock solution: 0.1M - 50mL
Made ampicillin stock solution: -1 g ampicillin -5mL DIwater -ethanol
Inoculated E. coli DH5α from frozen stock into 12 mL LB broth
~1.5 hrs 7/8/2010Alex, Jennifer and Eric - in ERB Made LB agar plates w/ ampicillin -10 g LB broth -7.5 g agar -500 mL DI water
Created protocols for Obtaining Deionized Water in the ERB and ERB Spectrophotometer
poured 20 LB-amp plates (large)
~3.5 hrs work Ann, Marc, Audra and Katie - in Lin Lab Performed biobrick transformations using heat shock
After second washing:
4 hrs of work 7/17/2010Alex & Jennifer - in ERB Yesterday, Marcus stored newly obtained strains in 4°C in ERB
Removed these strains from 4°C -- they are stored in soft agar stab cultures Made one streak plate on LB+amp for each strain from stabs
Made one 2mL LB+amp broth cultures in a 15mL tube for each strain from stabs
Placed agar stabs and LB+amp broth in 4°C ~1.5 hrs work Created protocol/plan for experiment. Testing AI-2 Response 7/19/2010Alex and Marcus 7/17 plates are no good -- they needed kanamycin
Made LB agar -20 g powder/500 mL DI water
Obtained a rotor for 1.5mL tubes for the centrifuge from Rodger Pinto
Obtained LuxS (strain JW2662-1) & MarC (for Jeremy Minty) null mutant E. coli strains from [http://cgsc.biology.yale.edu/ CGSC]
Transfered iGEM cryobox from Lin Lab -20°C to ERB -20°C ~6 hrs 7/21/2010Alex, Eric and Jeremy {Yesterday, Marcus made broths for E. coli JW2662-1 (LuxS-) (from spread plate), E. coli JW1522-1 (MarC- for Jeremy Minty) (from spread plate), E. coli W3110 (from stab), E. coli MDAI2 (from stab), P. putida for Oil Sands and P. fluorescens for Oil Sands
Crystored all six strains following Making frozen stocks protocol
Plated LuxS- and MarC- mutants and Pseudomonas strains on LB Plated W3110 and MDAI2 on LB+amp+kan
Stored remaining clean LB+amp (6) and LB+amp+kan (8) plates from 7/19 in 4°C ERB 1239 ~1.5 hrs 7/22/2010Alex During Lab Committee meeting -- Moved the six plates from yesterday from 37°C to 4°C Uploaded Protocol: Culturing CGSC Strains
7/23/2010Alex Updated Strain Database 7/25/2010Alex and Eric Made 250 mL LB broth in each of two 500mL flasks -5 g LB broth -250 mL DI water
Autovclaved 250 mL DI water in each of two 500mL flasks
Made a 2mL culture in 15mL tube of W3110 -2 mL LB from 4°C -2 μL 100mg/mL ampicillin (final conc. 100μg/ml) -2 μL 50mg/mL kanamycin (final conc. 50μg/mL)
Left stuff in autoclave 7/26/2010Alex and Marcus {yesterday, Josh and Charlie removed stuff from the autoclave} Analyzed yesterday's W3110 culture on Lin microplate reader
Autoclaved two 500mL flasks to be sterile containers, each with ~150 mL DI water inside
Alex Learned how to use Epifluorescence Microscope in HHDow (2nd floor) from Alissa
Removed glassware from the autoclave --> ERB 1230 Made broth cultures for W3110 and MDAI2
7/27/2010Alex, Eric and Marcus Checked OD600 of yesterday's cultures in Lin Lab spectrometer
Need to start culture with OD .02
Obtained 40% glucose solution from Ann - Lin Lab
Started 100mL culture of W3110 in 500mL flask -2.29 mL W3110 culture -2 mL 40% glucose -95.7 mL LB
Started 100mL culture of MDAI2 in 500mL flask -2.34 mL MDAI2 culture -2 mL 40% glucose -95.7 mL LB
Marcus and Eric Alex and Marcus Checked remaining MDAI2 and W3110 cultures on fluroescence microscope in LSI 6th floor
Alex Read W3110 & MDAI2 cultures on microplate reader in Xi Lab - SPH
7/28/2010Alex Made 40 mL of 0.1% crystal violet solution in Xi Lab - SPH
7/30/2010Alex and Marcus - w/ Charlie and Prae Performed transformation of 5 parts following Transformation-electroporation protocol -- starting from reading OD of overnight. (Ann started the previous steps.)
Continued with protocol...
8/2/2010Alex and Eric Read overnight cultures on Lin Lab spectrometer
OK, I don't know if these have GFP or what. At any rate, if they do, the MDAI2 is too close to W3110. We'll probably have to work with the YFP biobrick instead. Got C. vulgaris from Bobby Levine
Alex and Marcus Made broth cultures in 15mL tubes from plates
Autoclaved waste flasks
8/3/2010Alex and Marcus Took yesterday's overnights to Alex's (Xi) lab since the Lin lab fluorospectrophotometer was in use.
Fluorescence/OD600:
Very confusing results, LuxS- was supposed to be the negative control, and shows the highest fluorescence per OD. Some factor must be confounding our assay. Also, made 10 mL overnights of LuxS- (pLsr-YFP) and MDAI2 (pET6 + pET-GFP) in 50 mL tubes at 7 pm in LB+Amp+Kan. Put in shaker at 30°C. 8/4/2010Alex and Marcus Made broths for experiment tomorrow
Made stocks of LB + 50μg/mL kan (25 mL) and of LB + 100μg/mL amp + 50μg/mL kan (50 mL)
Updated QS experimental protocol
8/5/2010Alex Continued testing AI-2 response of yesterday's cultures, following Lsr Circuit Test Protocols
Followed protocol
Continued protocol...
Continued Protocol
Attended Lab Committee meeting Researched Heidelberg team uploaded notebook made presentation for tomorrow Finished exp by protocol
8/19/2010Alex Acquired new strains (soft agar stabs from Tsao Lab)
Made broth cultures of MDAI2 and BL21 from stab cultures
Added Quorum Sensing project description to Wiki 8/20/2010Alex [Earlier, Marcus cryostored BL21 and MDAI2 in Lin -80°C and made a spread plate of new MDAI2 strain.] Made streak plate of BL21
Made frozen stock of BL21 and MDAI2
Updated strain database 8/22/2010Alex Moved MDAI2 spread plate and BL21 streak plate from 37°C to 4°C Made BL21 broth culture
8/23/2010Marcus Measured OD600 of overnight in Gulari Lab spectrophotometer. -Used LB+ Amp(100μg/mL)+ Kan(50μg/mL) blank -OD was 1.55 Made dilution to OD 0.02 into 50 mL flask using OD600 Cell Dilution Protocol. -48.3 mL LB -1 mL 40% glucose solution -0.645 mL overnight culture Put in 37°C shaker for 4 hours and then collected supernatant according to Lsr Circuit Test Protocols. Supernatant was labeled and stored in the ERB -20°C. 8/26/2010Alex and Marcus Made cultures of MDAI2 + pTC6 + pET-GFP (new) and LuxS- + pLsrA-YFP
8/27/2010Alex Obtained cultures and supernatants from ERB -- 1:15pm (16 hrs); transfered to Xi Lab, SPH Began Quorum Sensing experiment again, following posted protocol
Returned supernatants to ERB -20°C 8/28/2010Alex Obtained data from QS main experiment
Only the wells with LB added to pellet had a significant increase in fluorescence over time.
It seems that the LB blank well was contaminated slightly. This shouldn't really matter, unless the contamination was also in the other two LB wells, in which case maybe co-culture was what cause the increase in fluorescence. However, OD also only increased significantly for the LB wells, so this is probably why the fluorescence also increased. This does present another experiment idea, however: maybe we can try to culture MDAI2 alone, and then in co-culture with K12 to see if there it a difference in GFP. At any rate, I'm not sure it's feasible to co-culture with algae, so quorum sensing project is probably done. |
In the Lab |
 "
"