Team:VT Ensimag 2010 Biosecurity/DNAkeep
From 2010.igem.org
(Difference between revisions)
(New page: {{Template:Team:VT_Ensimag_2010-Biosecurity/Templates/subpage|Nucleotide| content= __NOTOC__ We have to screen both nucleotide and amino-acid sequences according to the federal guideline. ...) |
|||
| (One intermediate revision not shown) | |||
| Line 4: | Line 4: | ||
For example, the complete genome of Mycoplasma mycoides subsp. mycoides SC str. PG1 has an entry only for nucleotide sequence, and any for amino-acid. | For example, the complete genome of Mycoplasma mycoides subsp. mycoides SC str. PG1 has an entry only for nucleotide sequence, and any for amino-acid. | ||
| - | [[Image:VTENSI_Exgenbank.png|center| | + | [[Image:VTENSI_Exgenbank.png|center|600px]] |
<br> | <br> | ||
<br> | <br> | ||
| - | [[VT-ENSIMAG/Genothreat|Go back to genothreat page]] | + | [[Team:VT-ENSIMAG/Genothreat|Go back to genothreat page]] |
}} | }} | ||
Latest revision as of 21:10, 9 August 2010
Nucleotide
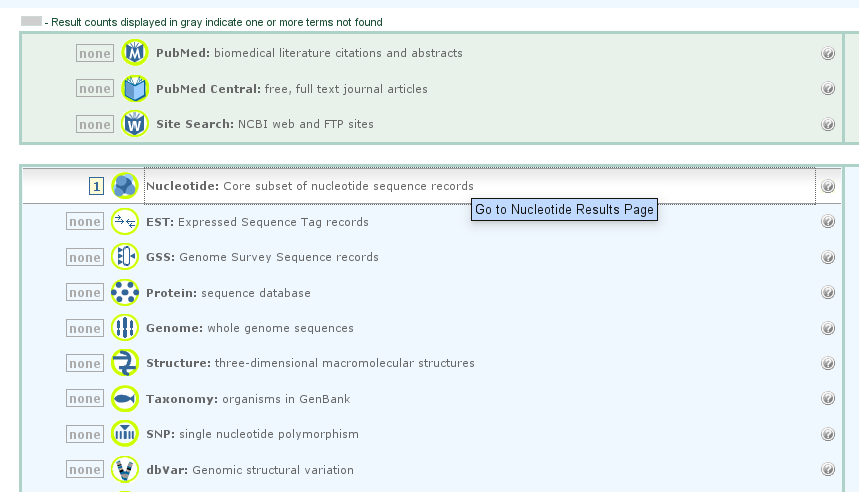
We have to screen both nucleotide and amino-acid sequences according to the federal guideline. Indeed, one nucleotide sequence can create 6 different polypeptides, so we have to look at each one. Moreover, the Genbank database doesn't obligatory contain both nucleotide and amino-acid sequence for one element. So we have to screen also the nucleotide sequence.
For example, the complete genome of Mycoplasma mycoides subsp. mycoides SC str. PG1 has an entry only for nucleotide sequence, and any for amino-acid.
 "
"