|
AUGUST: WEEK 1
August, 2nd
Miniprep and quantification with Nanodrop of:
- I20-1: 98,2 ng/ul
- I20-2: 63,6 ng/ul
- I20-3: 41,5 ng/ul
- I21-1: 45 ng/ul
- I21-2: 45 ng/ul
- I21-3: 54 ng/ul
These samples were prepared and sent (400ng) to BMR Genomics for sequencing.
The following parts were resuspended from iGEM 2010 Distribution Kit:
- <partinfo>BBa_R0062</partinfo> (Plate 1, Well 6O)
- <partinfo>BBa_K081009</partinfo> (Plate 2, Well 10N)
both in vector <partinfo>pSB1A2</partinfo>.
Transformation (1ul) of the following parts (resuspended/already miniprepped):
| Part | Strain | Culture name
|
| pAH123 | MC1061 | MC123
|
| MG1655 | MG123
|
| <partinfo>BBa_J72008</partinfo> | MC1061 | MC008
|
| MG1655 | MG008
|
| <partinfo>BBa_R0062</partinfo> | DH5-alpha |
|
| <partinfo>BBa_K081009</partinfo>
|
Transformed cells were plated on proper LB+Amp agar plates and grown ON at right temperature:
| Part | Plate resistance | Temperature
|
| pAH123 | Amp 50 ug/ml | 30°C
|
| <partinfo>BBa_J72008</partinfo>
|
| <partinfo>BBa_R0062</partinfo> | Amp 100 ug/ml | 37°C
|
| <partinfo>BBa_K081009</partinfo>
|
August, 3rd
Check for plates grown ON: all plates showed colonies.
 <partinfo>BBa_R0062</partinfo> plate |  <partinfo>BBa_K081009</partinfo> plate |
A single colony was picked from <partinfo>BBa_R0062</partinfo> and <partinfo>BBa_K081009</partinfo> plates and inoculated in 1 ml LB+Amp 100 ug/ml and incubated 37°C, 220 rmp for glycerol stock. Cultures left were refilled to 5 ml of proper medium and incubated ON at 37°C, 220 rpm for further screening.
Since plates left showed small colonies they were let grow until late afternoon; than some colonies were picked from each plate and inoculated into a 5 ml LB+Amp 50 ug/ml falcon. Falcon tubes were incubated and shaken ON at 30°C.
 MC1061 transformed with pAH123 |  MC1061 transformed with <partinfo>BBa_J72008</partinfo> |
 MG1655 transformed with pAH123 |  MG1655 transformed with <partinfo>BBa_J72008</partinfo> |
PCR from the following colonies (this is a test for the efficiency of our primers synthesized to check attPhi80 E. coli genomic integration and it will be our negative control for future screenings):
- MC1061-1
- MC1061-2
- MG1655-1
- MG1655-2
- Blank (Nothing)
Gel run of amplified DNA showed for every sample the expected distance between primers of a strain with nothing integrated in attPhi80 site (~570 bp), but unfortunately we forgot to take a picture of the gel ;(
August, 4th
Glycerol stocks for MC1061 and MG1655 strains transformed with pAH123 or <partinfo>BBa_J72008</partinfo> helper plasmids.
2 ul of MC1061 bacteria were transferred into 5 ml LB+Amp 50 ug/ml and grown and shaken at 30°C over-day and over-night for re-competentization of the following day.
200 ul of MG1655 cultures were transferred into 100 ml LB+Amp 50 ug/ml and grown and shaken at 30°C for re-competentization of today.
All cultures were miniprepped to check again the presence of helper plasmids.
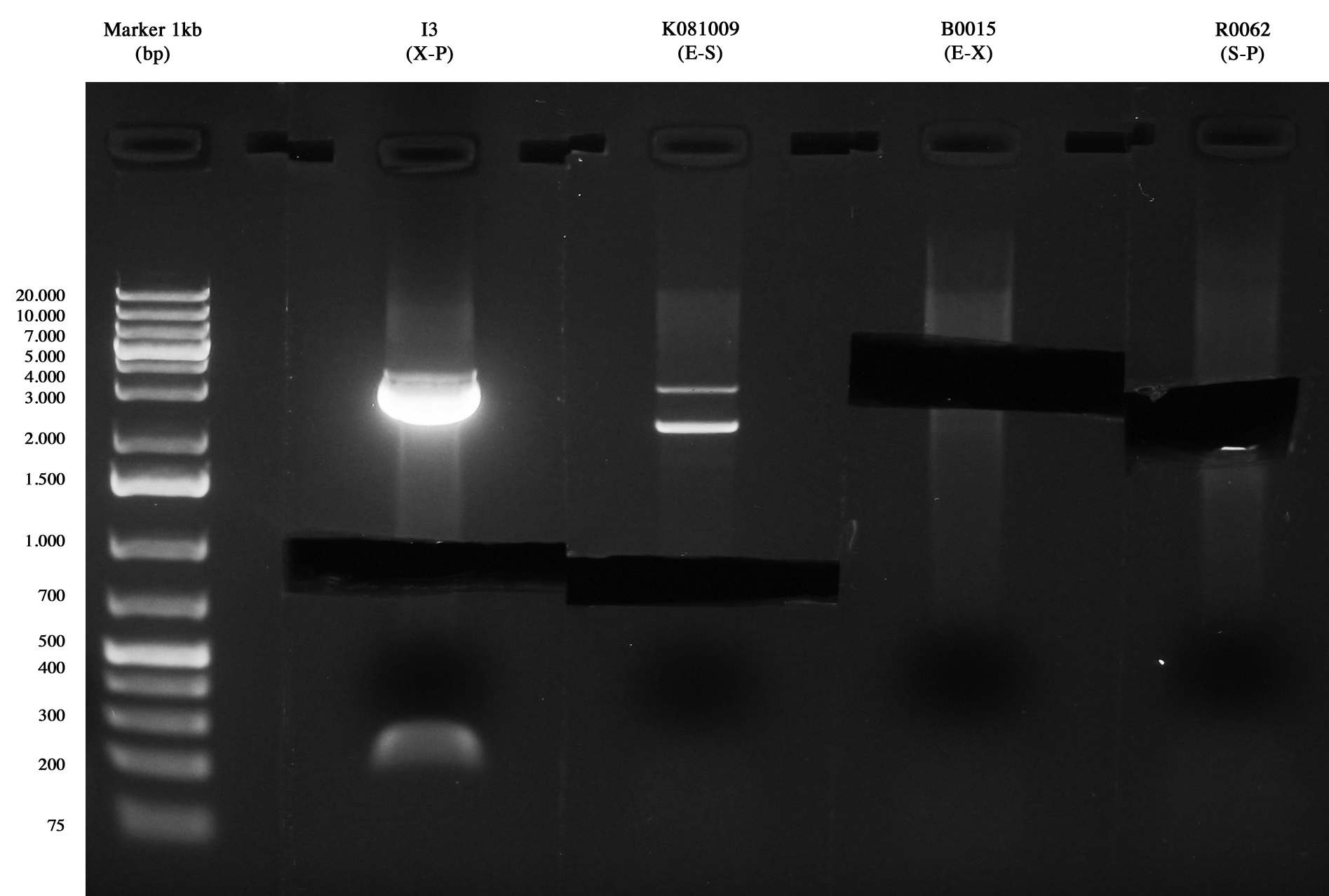
Samples were digested with SpeI (it cuts twice) for 3 hours and gel run:
- pAH123 digested: 3580 and 2755 bp
- <partinfo>BBa_J72008</partinfo> digested: 2755 and 2437 bp
 pAH123 and <partinfo>BBa_J72008</partinfo> screening transformed into MG1655 and MC1061 Samples are positive (right lengths) but unfortunately we got a bad gel run (smearings) so this time we decided to pick two single colonies from each of the plates made on August, 3rd and to inoculate them into 5 ml LB+Amp 50 ug/ml. A total of eight falcon tubes was incubated ON at 30°C, 220 rpm.
We planned to screen them (to check the presence of helper plasmids again) and to re-competentize only the positive ones.
August, 5th
Glycerol stock and miniprep of MG1655 and MC1061 cultures incubated for 19 hours at 30°C, 220 rpm.
Miniprep was quantified as follows:
- MG123-1: 27 ng/ul
- MG123-2: 40,2 ng/ul
- MG008-1: 20,5 ng/ul
- MG008-2: 26,5 ng/ul
- MC123-1: 21,4 ng/ul
- MC123-2: 17,4 ng/ul
- MC008-1: 33,3 ng/ul
- MC008-2: 38,1 ng/ul
DIgestion for 1 hour with SpeI.
Gel run on medium agarose gel.
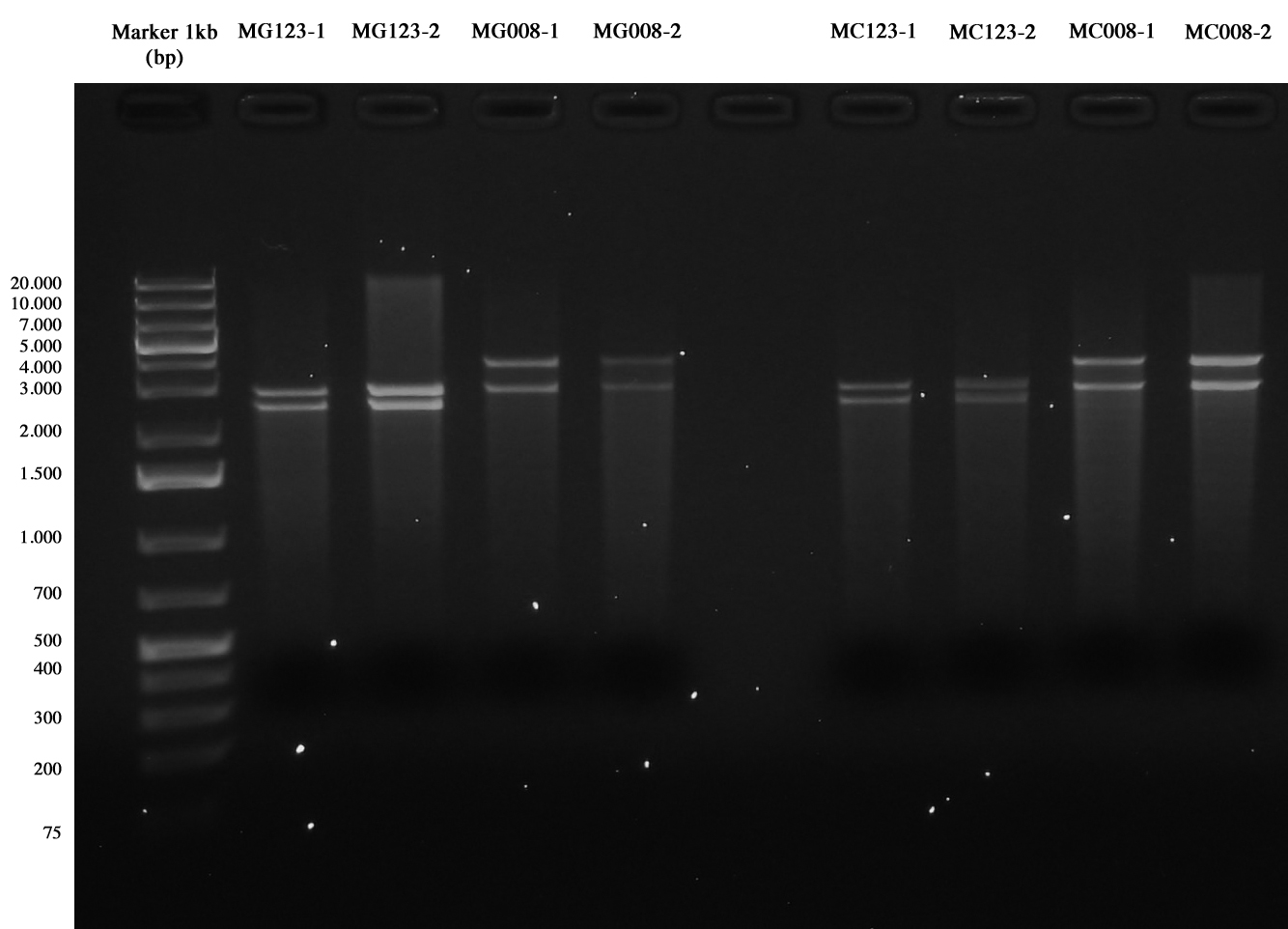 pAH123 and <partinfo>BBa_J72008</partinfo> screening (miniprepped from MG1655 and MC1061) This time gel run succeeded, we chose samples
- MG123-1
- MG008-1
- MC123-1
- MC008-1
to be re-competentizied. So they were inoculated into 5 ml LB+Amp 50 ug/ml and grown and shaken ON at 30°C.
Sequencing for I20 and I21 arrived from BMR, but all samples were wrong; sites X and S of the vector paired and nothing could ligate. So we started a new ligation cycle dephosphorylating the previously gel-extracted vector <partinfo>pSB1A3</partinfo>.
New ligations:
- I20-new: Pha-10S-1 (X-S) + <partinfo>pSB1A3</partinfo> (X-S, dephosphorylated)
- I21-new: Pha-SS-1 (X-S) + <partinfo>pSB1A3</partinfo> (X-S, dephosphorylated)
and their negative control:
- C-: <partinfo>pSB1A3</partinfo> (X-S, dephosphorylated)
August, 6th
Resuspension of linker <partinfo>BBa_K105012</partinfo> from iGEM 2010 Distribution Kit.
Transformation of ligations and resuspended DNA
- I20-new
- I21-new
- C-
- <partinfo>BBa_K105012</partinfo>
into 100ul E. coli DH5-alpha.
They were plated on LB+Amp 100ug/ml agar plates
Competentization of colonies selected the previous day:
- MG123 (without '-1' from now on)
- MG008
- MC123
- MC008
August, 7th
Plates of transformed cells were stored at +4°C.
|
 "
"