Team:UNIPV-Pavia/Calendar/July/settimana4
From 2010.igem.org
(→July, 22nd) |
m (→July, 21st) |
||
| (27 intermediate revisions not shown) | |||
| Line 32: | Line 32: | ||
<br> | <br> | ||
<html><p align="center"><font size="4"><b>JULY: WEEK 4</b></font></p></html><hr><br> | <html><p align="center"><font size="4"><b>JULY: WEEK 4</b></font></p></html><hr><br> | ||
| + | <html><a name="indice"/></html> | ||
==July, 19th== | ==July, 19th== | ||
| Line 74: | Line 75: | ||
BW23473 arrived from Yale University on a paper disk. It was grown ON in 5ml L, at 37°C, 220 rpm. | BW23473 arrived from Yale University on a paper disk. It was grown ON in 5ml L, at 37°C, 220 rpm. | ||
| + | |||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==July, 20th== | ==July, 20th== | ||
| Line 210: | Line 213: | ||
Inoculum of MC1061 in LB, PBHR68 in LB+Amp and <partinfo>BBa_T9002</partinfo> in LB+Amp to prepare competent cells tomorrow. | Inoculum of MC1061 in LB, PBHR68 in LB+Amp and <partinfo>BBa_T9002</partinfo> in LB+Amp to prepare competent cells tomorrow. | ||
| - | + | <div align="right"><small>[[#indice|^top]]</small></div> | |
| - | < | + | |
| + | ==July, 21st== | ||
This morning all cultures were grown: | This morning all cultures were grown: | ||
*BW25141-RING | *BW25141-RING | ||
| Line 330: | Line 333: | ||
</table> | </table> | ||
| - | Plates were incubated overnight at 37°C | + | Plates were incubated overnight at 37°C. |
| + | |||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==July, 22nd== | ==July, 22nd== | ||
| - | + | Plates results were not nice: I15 new showed few colonies (about 20 colonies), I14_4C5 showed 2 colonies, I16_4C5 showed 3 colonies and I18_4C5 1 colony, while for other plates no colony was observed, probably due to the fact that our 4C5 (E-P) plasmid retrieved from freezer was too old and not usable anymore. For this reason, we decided to perform a colony PCR on available colonies of I14_4C5, I16_4C5 and I18-4C5, on 3 colonies of I15new and to repeat wrong ligations and the one including I10-1 and I12-2 (their vector had to be changed with pSB4C5). | |
Colonies were saved in 1ml LB+suitable antibiotic/s. After gel results positive colonies will be glycerol stocked. | Colonies were saved in 1ml LB+suitable antibiotic/s. After gel results positive colonies will be glycerol stocked. | ||
| - | [[Image:UNIPV10_colony_pcr_I15new_I14-18.jpg|thumb|200px|center|Colony PCR of colonies | + | [[Image:UNIPV10_colony_pcr_I15new_I14-18.jpg|thumb|200px|center|Colony PCR of colonies picked from plates]] |
Colony PCR and gel run showed that I15-1, I15-2 and I15-3 have the right insert. Also for I14_4C5-1 and I14_4C5-2 the insert was visible. For these parts we decided to perform a NheI-PstI screening. I16-4C5-1 and I16_4C5-3 showed the insert, so they will be screened as before. No band was observed for I16_4C5-2 and I18_4C5-1 (This ligation will be repeated). | Colony PCR and gel run showed that I15-1, I15-2 and I15-3 have the right insert. Also for I14_4C5-1 and I14_4C5-2 the insert was visible. For these parts we decided to perform a NheI-PstI screening. I16-4C5-1 and I16_4C5-3 showed the insert, so they will be screened as before. No band was observed for I16_4C5-2 and I18_4C5-1 (This ligation will be repeated). | ||
| Line 363: | Line 368: | ||
We also performed a TECAN test in order to evaluate the ranking of sternght of promoters we inoculaed yesterday. Results how that the ranking is the one reported here (from stronger to weaker): | We also performed a TECAN test in order to evaluate the ranking of sternght of promoters we inoculaed yesterday. Results how that the ranking is the one reported here (from stronger to weaker): | ||
| - | *< | + | *<partinfo>BBa_J23100</partinfo> |
| - | *< | + | *<partinfo>BBa_J23101</partinfo> |
| - | *< | + | *<partinfo>BBa_J23110</partinfo> |
| - | *< | + | *<partinfo>BBa_J23118</partinfo> |
| - | *< | + | *<partinfo>BBa_J23106</partinfo> |
| - | *< | + | *<partinfo>BBa_J23105</partinfo> |
| - | *< | + | *<partinfo>BBa_J23116</partinfo> |
| - | *< | + | *<partinfo>BBa_J23114</partinfo> |
| + | |||
| + | Today we also prepared new plates, because we finished the one prepared, for our intensive lab activity... | ||
| + | 50 plates LB+Amp+Cm 12,5. | ||
| + | |||
| + | We transformed pSB4C5 (4ng) in our home made competent cells: | ||
| + | *T9002 | ||
| + | *MC1061 | ||
| + | *PBHR68 | ||
| + | to evaluate transformation efficiency. | ||
| - | + | <div align="right"><small>[[#indice|^top]]</small></div> | |
| - | + | ||
==July, 23rd== | ==July, 23rd== | ||
| + | |||
| + | All plates incubated yesterday (T9002-4C5, MC1051-4C5 and PBHR68-4C5) showed colonies! We decided to incubate T9002 for furthrt time, because colonies were too small, before counting... Soon we will count colonies to evaluate efficiency of transformation for our home made competent cells! | ||
| + | |||
| + | |||
| + | Today we transformed our ligations: | ||
| + | |||
| + | <table width='90%' border='1'> | ||
| + | <tr><th>Ligation name</th><th>E. coli strain </th><th> Resistance </th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> | ||
| + | *I17_4C5= I17 (E-P) + pSB4C5 (E-P) | ||
| + | </td> | ||
| + | <td> | ||
| + | DH5alpha | ||
| + | </td> | ||
| + | <td>Cm 12,5</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> | ||
| + | *I18_4C5= I18 (E-P) + pSB4C5 (E-P) | ||
| + | </td> | ||
| + | <td> | ||
| + | DH5alpha | ||
| + | </td> | ||
| + | <td>Cm 12,5</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> | ||
| + | *I19_4C5= I19 (E-P) + pSB4C5 (E-P)</td> | ||
| + | <td> | ||
| + | DH5alpha | ||
| + | </td> | ||
| + | <td>Cm 12,5</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> | ||
| + | *I10_4C5= I0 (E-P) + pSB4C5 (E-P)</td> | ||
| + | <td> | ||
| + | TOP10 | ||
| + | </td> | ||
| + | <td>Cm 12,5</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> | ||
| + | *I12_4C5= I12 (E-P) + pSB4C5 (E-P)</td> | ||
| + | <td> | ||
| + | TOP10 | ||
| + | </td> | ||
| + | <td>Cm 12,5</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> | ||
| + | *Entero 4C5</td> | ||
| + | <td> | ||
| + | PBHR68 | ||
| + | </td> | ||
| + | <td>Amp 100+Cm 12,5</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> | ||
| + | *RING</td> | ||
| + | <td> | ||
| + | MC1061 | ||
| + | </td> | ||
| + | <td>Cm 12,5</td> | ||
| + | </tr> | ||
| + | <tr><td> | ||
| + | *No DNA (negative control)</td> | ||
| + | <td> | ||
| + | MC1061 | ||
| + | </td> | ||
| + | <td>Cm 12,5</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | |||
| + | |||
| + | |||
| + | After MiniPrep, purified DNA was quantified with NanoDrop. | ||
| + | |||
| + | {| border='1' align='center' | ||
| + | | '''Culture''' || '''Quantification''' | ||
| + | |- | ||
| + | | I15-1 || 34,2 ng/ul | ||
| + | |- | ||
| + | | I15-2 || 57,1 ng/ul | ||
| + | |- | ||
| + | | I15-3 || 72 ng/ul | ||
| + | |- | ||
| + | | I14_4C5-1 || 14,7 ng/ul | ||
| + | |- | ||
| + | | I14_4C5-2 || 20,9 ng/ul | ||
| + | |- | ||
| + | | I16_4C5-1 || 16,3 ng/ul | ||
| + | |- | ||
| + | | I16_4C5-2 || 15,3 ng/ul | ||
| + | |} | ||
| + | |||
| + | Digestion of: | ||
| + | |||
| + | {| border='1' | ||
| + | | ''Culture'' || ''Kind'' || ''Final reaction volume (ul) '' || ''DNA (ul)'' || ''H20 (ul)'' || ''Enzyme 1'' || ''Enzyme 2'' || ''Buffer B'' | ||
| + | |- | ||
| + | | I15-1 || Screening || 25 || 3 || 18,5 || 0,5 NheI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I15-2 || Screening || 25 || 3 || 18,5 || 0,5 NheI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I15-3 || Screening || 25 || 3 || 18,5 || 0,5 NheI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I14_4C5-1 || Screening || 25 || 10 || 11,5 || 0,5 NheI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I14_4C5-2 || Screening || 25 || 10 || 11,5 || 0,5 NheI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I16_4C5-1 || Screening || 25 || 10 || 11,5 || 0,5 NheI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I16_4C5-2 || Screening || 25 || 10 || 11,5 || 0,5 NheI || 0,5 PstI || 2,5 | ||
| + | |} | ||
| + | |||
| + | Digestions were incubated for 3h at 37°C, then gel run. | ||
| + | |||
| + | [[Image:UNIPV10_I15_I144C%_I164C5_screening.jpg|thumb|200px|center|Screening for I15, I14-4C5 and I16_4C5]] | ||
| + | |||
| + | Gel showed that all parts were ok!! | ||
| + | |||
| + | We also received sequencing results: | ||
| + | <html><b> | ||
| + | <table align='center' border='1' width='35%'> | ||
| + | <tr><td> | ||
| + | I14-1</td><td><font color='#009966'> | ||
| + | correct!</font></td></tr> | ||
| + | <tr><td> | ||
| + | I16-1</td><td><font color='#009966'> | ||
| + | correct!</font></td></tr> | ||
| + | <tr><td> | ||
| + | I17-1</td><td><font color='#009966'> | ||
| + | correct!</font></td></tr> | ||
| + | <tr><td> | ||
| + | I18-1</td><td><font color='#009966'> | ||
| + | correct!</font></td></tr> | ||
| + | <tr><td> | ||
| + | I19-1</td><td><font color='#009966'> | ||
| + | correct!</font></td></tr> | ||
| + | <tr><td> | ||
| + | I12-2</td><td><font color='#009966'> | ||
| + | correct!</font></td></tr> | ||
| + | <tr><td> | ||
| + | I7_4C5-2</td><td><font color='#009966'> | ||
| + | correct!</font></td></tr> | ||
| + | <tr><td> | ||
| + | I8_4C5-2</td><td><font color='#009966'> | ||
| + | correct!</font></td></tr> | ||
| + | </table></b> | ||
| + | </html> | ||
| + | |||
| + | Our plate with MC1061 transformed with RING showed few contamination, so we decided to repeat the transformation. | ||
| + | |||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==July, 24th== | ==July, 24th== | ||
| + | |||
| + | We retransformed RING in MC1061 in order to see if contamination we noticed yesterday was due to tranformation or not. Plate was incubated overnight at 37°C and tomorrow we will evaluate efficiency. | ||
| + | |||
| + | For plates incubated yesterday, we picked 2 colonies for each plate: | ||
| + | *I10_4C5-1 | ||
| + | *I10_4C5-2 | ||
| + | *I12_4C5-1 | ||
| + | *I12_4C5-2 | ||
| + | *I17_4C5-1 | ||
| + | *I17_4C5-2 | ||
| + | *I18_4C5-1 | ||
| + | *I18_4C5-2 | ||
| + | *I19_4C5-1 | ||
| + | *I19_4C5-2 | ||
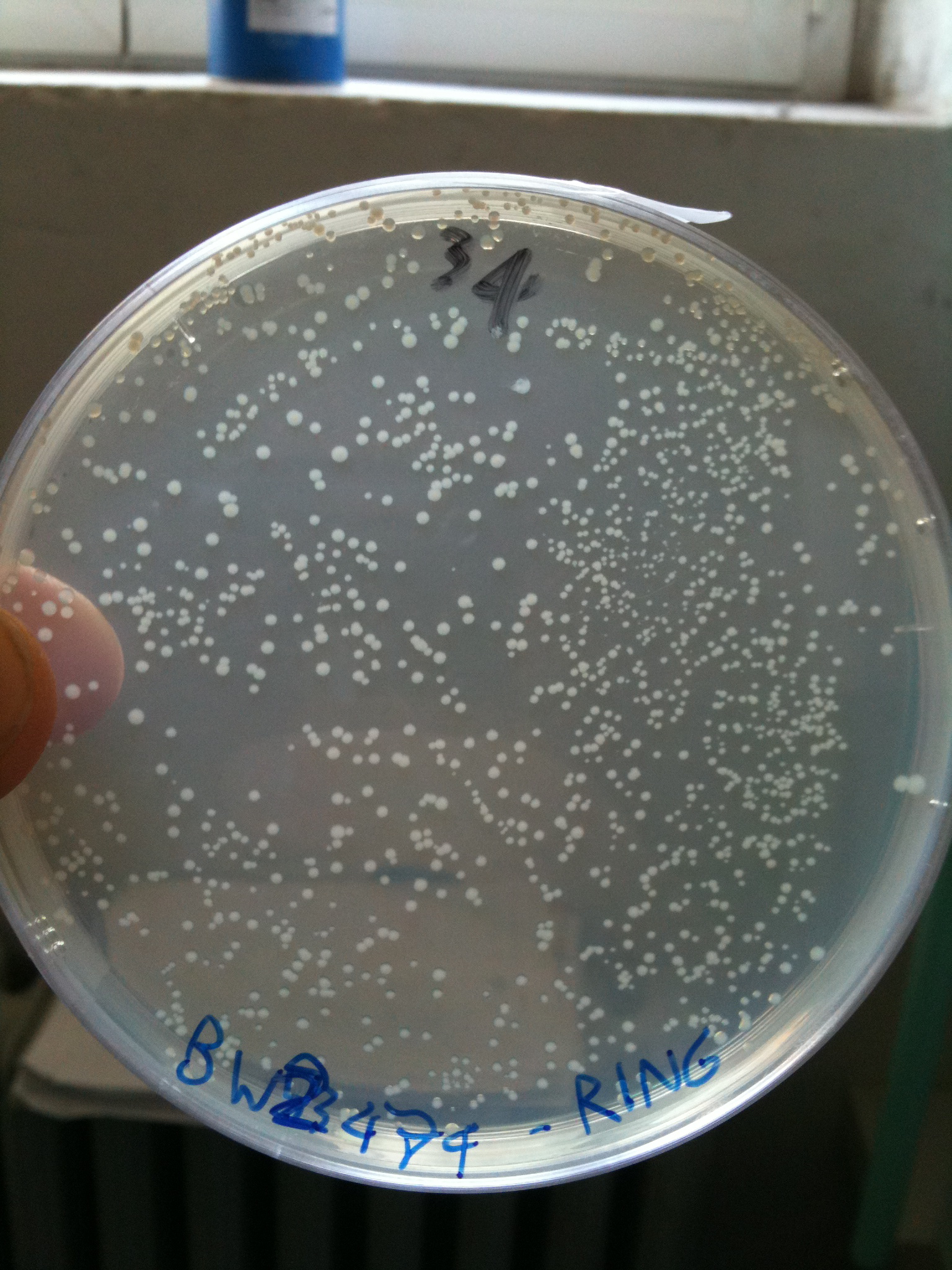
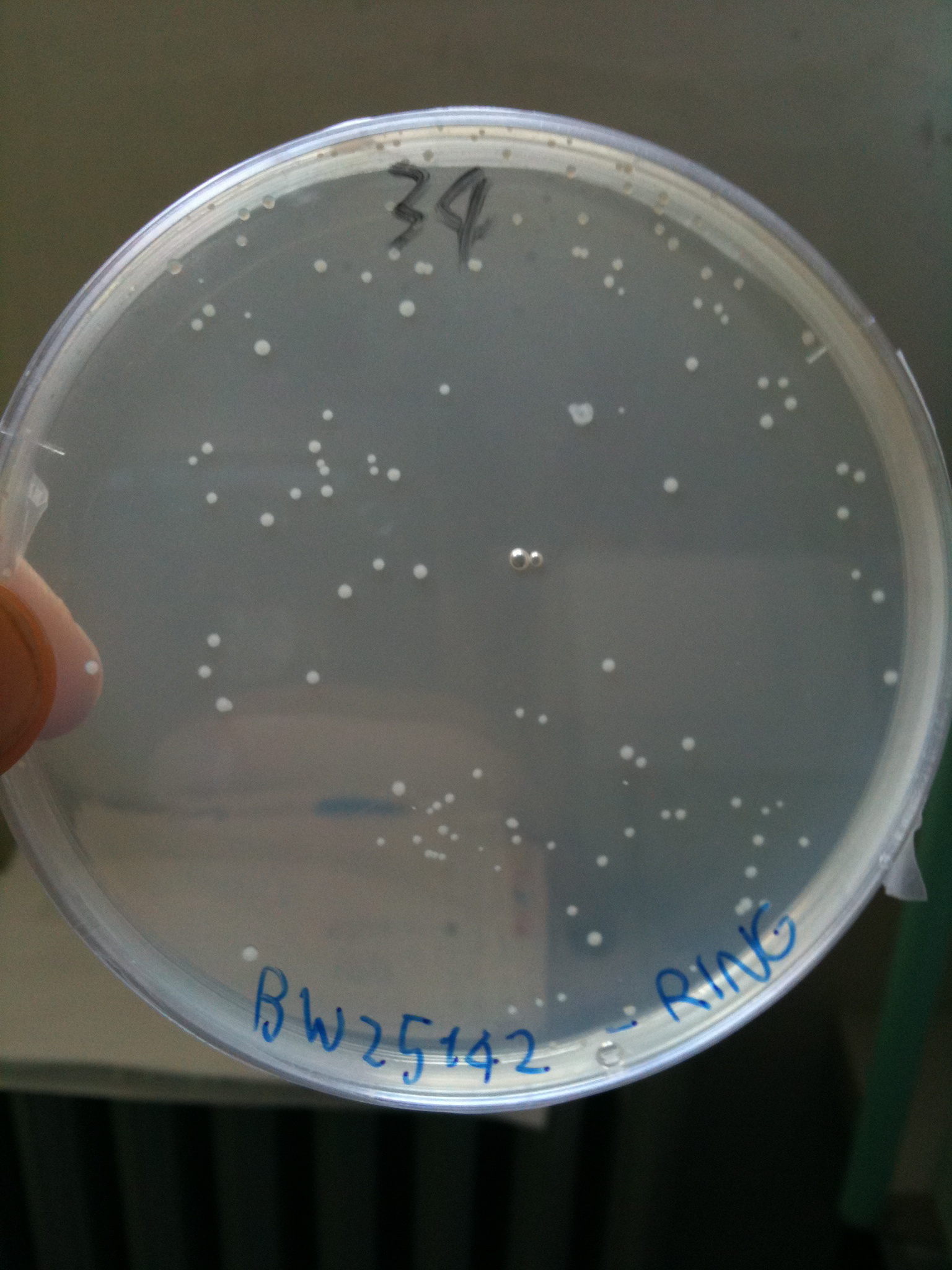
Efficiency of transformation: | Efficiency of transformation: | ||
| Line 390: | Line 574: | ||
|BW23474 || 1436 colonies | |BW23474 || 1436 colonies | ||
|} | |} | ||
| + | |||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==July, 25th== | ==July, 25th== | ||
| + | The plate containing RING in MC1061 showed 3 colonies! These colonies were screened and grown in LB+Cm 12,5. For colonies picked yesterday screening digestion was performed. | ||
| + | MiniPrep was performed for: | ||
| + | |||
| + | *I10_4C5-1 | ||
| + | *I10_4C5-1 | ||
| + | *I12_4C5-1 | ||
| + | *I12_4C5-1 | ||
| + | *I17_4C5-1 | ||
| + | *I17_4C5-1 | ||
| + | *I18_4C5-1 | ||
| + | *I18_4C5-1 | ||
| + | *I19_4C5-1 | ||
| + | *I19_4C5-1 | ||
| + | |||
| + | Digestion of: | ||
| + | |||
| + | {| border='1' | ||
| + | | ''Culture'' || ''Kind'' || ''Final reaction volume (ul) '' || ''DNA (ul)'' || ''H20 (ul)'' || ''Enzyme 1'' || ''Enzyme 2'' || ''Buffer Tango'' | ||
| + | |- | ||
| + | | I10-4C5-1 || Screening || 25 || 10 || 11,5 || 0,5 NheI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I10-4C5-1 || Screening || 25 || 10 || 11,5 || 0,5 NheI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I12-4C5-1|| Screening || 25 || 10 || 11,5 || 0,5 NheI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I12_4C5-2 || Screening || 25 || 10 || 11,5 || 0,5 NheI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I17_4C5-1 || Screening || 25 || 10 || 11,5 || 0,5 NheI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I17_4C5-2 || Screening || 25 || 10 || 11,5 || 0,5 NheI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I18_4C5-1 || Screening || 25 || 10 || 11,5 || 0,5 NheI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I18_4C5-2 || Screening || 25 || 10 || 11,5 || 0,5 NheI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I19_4C5-1 || Screening || 25 || 10 || 11,5 || 0,5 NheI || 0,5 PstI || 2,5 | ||
| + | |- | ||
| + | | I19_4C5-2 || Screening || 25 || 10 || 11,5 || 0,5 NheI || 0,5 PstI || 2,5 | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
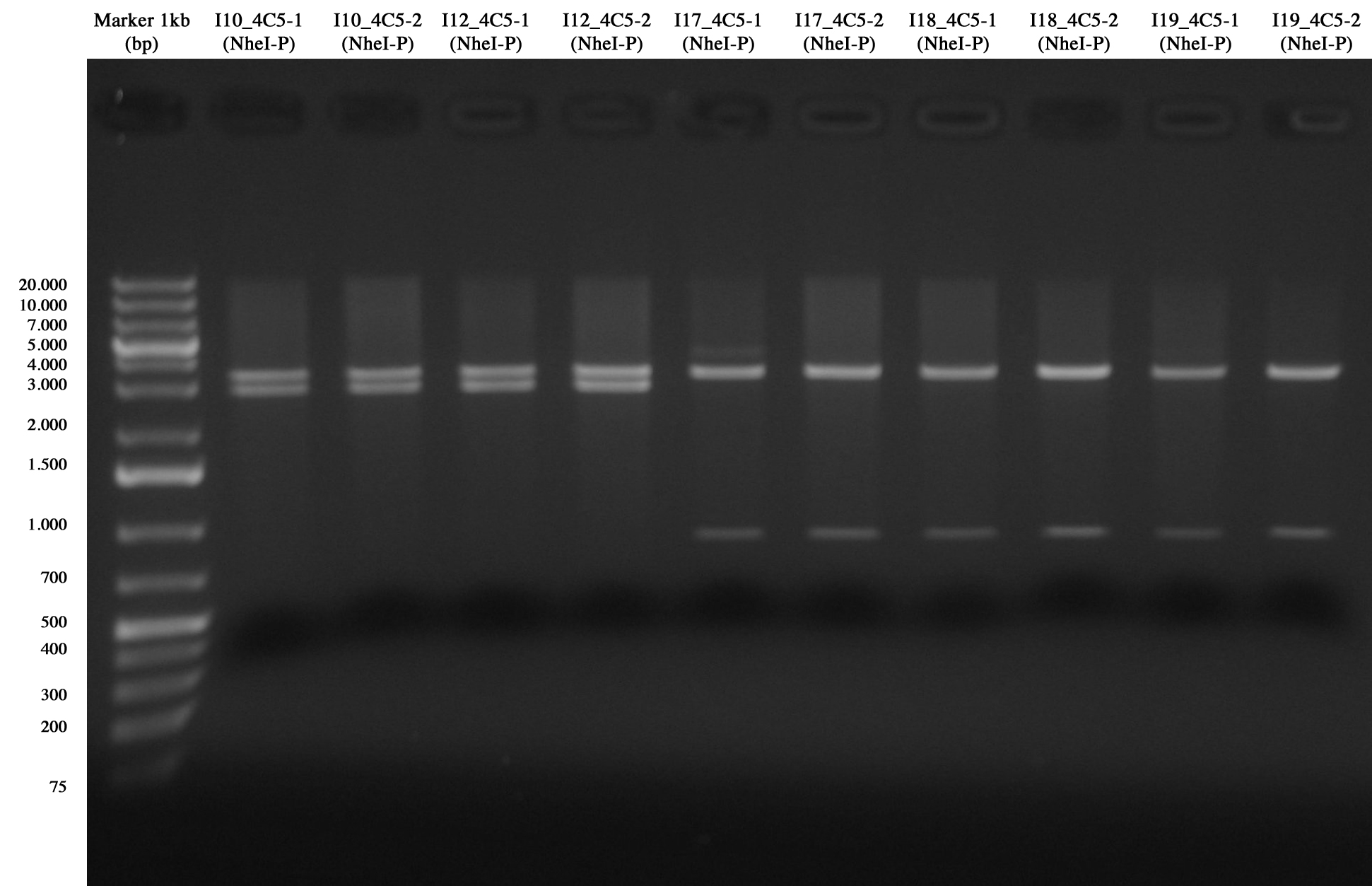
| + | [[Image:UNIPV10_II10_12_17_18_19_4C5_screening.jpg|thumb|200px|center|Screening for I10_4C5, I12_4C5, I17_4C5, I18_4C5 and I19_4C5]] | ||
| + | |||
| + | |||
| + | From gel it is possible to see that all of them are positive!! :) | ||
| + | |||
| + | |||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
| + | |||
<!-- table previous next week --> | <!-- table previous next week --> | ||
Latest revision as of 16:54, 24 October 2010
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"