Week of 6/28
From 2010.igem.org
(Difference between revisions)
(New page: <center>Image:6-29-10.jpg</center> <center>Image:6-30-10.jpg</center> <center>Image:6-30-10 001.jpg</center> <center>Image:6-1-10.jpg</center> <center>[[Image:6-2-1...) |
|||
| (11 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | ==June 28== | ||
| + | |||
| + | * Retransform: | ||
| + | -R0061 | ||
| + | -R0062 | ||
| + | -R0063 | ||
| + | -K091146 | ||
| + | |||
| + | *Inventory | ||
| + | -Good Sequencing: | ||
| + | o Backbone - C3RFP | ||
| + | o pLux - K091146 | ||
| + | o Lysis - K112022 | ||
| + | o pLux - R0063 | ||
| + | o pLux - R1062 | ||
| + | o CI+RBS - K081007 | ||
| + | o Cons.Prom- J23116 | ||
| + | o Cons.Prom- J23108 | ||
| + | o Pigment - K274100 | ||
| + | o LuxI - C0261 | ||
| + | o Lysis - K112808 | ||
| + | o pLac - R0010 | ||
| + | o LacI+RBS - J24679 | ||
| + | |||
| + | *To grow: | ||
| + | - K091146 | ||
| + | - R0063 | ||
| + | - J23108 | ||
| + | - J23116 | ||
| + | |||
| + | *Electrocompetent cells were made. | ||
| + | |||
| + | |||
| + | ==June 29== | ||
| + | |||
| + | Gel Order: | ||
| + | - J23108 | ||
| + | - J23108 (Digest 2) | ||
| + | - R0063 | ||
| + | - R0063 (Digest 2) | ||
| + | - K081007 | ||
| + | - K081007(Digest 2) | ||
| + | - J23116 | ||
| + | - J23116 (Digest 2) | ||
| + | |||
| + | |||
<center>[[Image:6-29-10.jpg]]</center> | <center>[[Image:6-29-10.jpg]]</center> | ||
| + | |||
| + | |||
| + | *Gel extract was done using Digest number 1. In this gel we ran the same part twice but from two different digests. Digest 1 was a 6 hour digest in the 37 degree Celsius water bath while Digest 2 was a 1 hour digest in the same water bath. As you can see, the results do not differ all that noticeably. That being established we went back to doing 1 hour digests. | ||
| + | |||
| + | *The bands that were cut out are provided below: | ||
| + | - J23108 | ||
| + | - R0063 | ||
| + | - J23116 | ||
| + | - K081007 | ||
| + | |||
| + | All these bands were purified. | ||
| + | |||
| + | ==June 30== | ||
| + | |||
| + | |||
| + | *On this gel, we ran the Colony PCR and the digests. The order is as follows: | ||
| + | |||
| + | *Colony PCR: | ||
| + | - Ladder | ||
| + | - A-J | ||
| + | |||
| + | *Digests: | ||
| + | - Ladder | ||
| + | - K091107 (digest 2) | ||
| + | - K091107 | ||
| + | - K091146 (digest 2) | ||
| + | - K091146 | ||
| + | - J23116+J37033 (digest 2) | ||
| + | - J23116+J37033 | ||
| + | - R0010 (digest 2) | ||
| + | - R0010 | ||
<center>[[Image:6-30-10.jpg]]</center> | <center>[[Image:6-30-10.jpg]]</center> | ||
| + | *The bands that were cut are marked with a square around them in the picture above. They were also then purified. | ||
| + | * 3 bands that were cut: | ||
| + | - K091107 | ||
| + | - K091146 | ||
| + | - R0010 | ||
| + | |||
| + | |||
| + | |||
| + | *A second gel was run the same day. The order is provided below: | ||
| + | - Ladder | ||
| + | - B1002 | ||
| + | - Lux R 2 | ||
| + | - C0261+J24679 | ||
<center>[[Image:6-30-10 001.jpg]]</center> | <center>[[Image:6-30-10 001.jpg]]</center> | ||
| + | |||
| + | *As you can see, the only 2 bands that were cut were B1002 and C0261+J24679 | ||
| + | |||
| + | |||
| + | *Grow for tomorrow: | ||
| + | - K274100 | ||
| + | - K112022 | ||
| + | |||
| + | ==July 1== | ||
| + | |||
| + | |||
| + | *Flasks minipreped: | ||
| + | - B1006 | ||
| + | - K274100 | ||
| + | - J23100 | ||
| + | |||
| + | *Digests: | ||
| + | - B1006 (EX) | ||
| + | - K274100 (XP) | ||
| + | - J23100 (SP) | ||
| + | |||
| + | *Sent to the sequencing center: | ||
| + | 1) B1002 | ||
| + | 2) B1006 | ||
| + | 3) S01414 | ||
| + | 4) J23100 | ||
| + | 5) C0261+J24679 | ||
| + | |||
| + | |||
| + | *Gel order: | ||
| + | - Ladder | ||
| + | - B1006 | ||
| + | - K274100 | ||
| + | - J23100 | ||
| + | |||
| + | <center>[[Image:7-1-10.jpg]]</center> | ||
| + | |||
| + | |||
| + | *All the bands appeared where we wanted them to so we cut out all three bands and then purified them. | ||
| + | |||
| + | |||
| + | |||
| + | *Ligations: | ||
| + | {| | ||
| + | |Vector||Inserts | ||
| + | |- | ||
| + | |1)J23100||K274100 | ||
| + | |- | ||
| + | |2)J23100||S01414 | ||
| + | |- | ||
| + | |3)R0063||a)K274100<br />b)C0261 + J24679 | ||
| + | |- | ||
| + | |4)K091146||a)K274100<br />b)C0261 + J24679 | ||
| + | |- | ||
| + | |5)K091107||a)K274100<br />b)C0261 + J24679 | ||
| + | |- | ||
| + | |6)R0062||a)K274100<br />b)C0261 + J24679 | ||
| + | |- | ||
| + | |7)R0010||K081007 | ||
| + | |} | ||
| + | |||
| + | *Numbers 3-6 were each matched up with a) K274100 and b) C0261 + J24679. The rest of the vectors were matched with the insert directly to the right in the table. | ||
| + | |||
| + | |||
| + | *Tomorrow: | ||
| + | - Develop primers to figure out if K274100 and K112808 are correct. | ||
| + | |||
| + | ==July 2== | ||
| + | |||
| + | |||
| + | *Colony PCRs run on gel.The order is as follows: | ||
| + | - Ladder | ||
| + | - 1A, 1B, 1C | ||
| + | - 2A, 2B, 2C | ||
| + | - 3A, 3B, 3C | ||
| + | |||
| + | - Ladder | ||
| + | - 4A, 4B, 4C | ||
| + | - 5A, 5B, 5C | ||
| + | - 6A, 6B, 6C | ||
| + | - 7A, 7B, 7C | ||
| + | |||
| + | <center>[[Image:7-2-10.jpg]]</center> | ||
| + | |||
| + | |||
| + | Unfortunately, we accidentally put ladder in all the Colony products. The picture is provided above. We could not use this information in any way other than to learn from our mistake. | ||
| - | |||
| - | + | [https://2010.igem.org/Team:Penn_State/Notebook Return to Notebook] | |
Latest revision as of 15:42, 5 August 2010
Contents |
June 28
- Retransform:
-R0061 -R0062 -R0063 -K091146
- Inventory
-Good Sequencing:
o Backbone - C3RFP
o pLux - K091146
o Lysis - K112022
o pLux - R0063
o pLux - R1062
o CI+RBS - K081007
o Cons.Prom- J23116
o Cons.Prom- J23108
o Pigment - K274100
o LuxI - C0261
o Lysis - K112808
o pLac - R0010
o LacI+RBS - J24679
- To grow:
- K091146 - R0063 - J23108 - J23116
- Electrocompetent cells were made.
June 29
Gel Order:
- J23108 - J23108 (Digest 2) - R0063 - R0063 (Digest 2) - K081007 - K081007(Digest 2) - J23116 - J23116 (Digest 2)
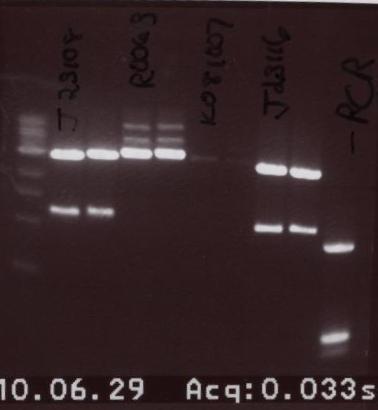
- Gel extract was done using Digest number 1. In this gel we ran the same part twice but from two different digests. Digest 1 was a 6 hour digest in the 37 degree Celsius water bath while Digest 2 was a 1 hour digest in the same water bath. As you can see, the results do not differ all that noticeably. That being established we went back to doing 1 hour digests.
- The bands that were cut out are provided below:
- J23108 - R0063 - J23116 - K081007
All these bands were purified.
June 30
- On this gel, we ran the Colony PCR and the digests. The order is as follows:
- Colony PCR:
- Ladder - A-J
- Digests:
- Ladder - K091107 (digest 2) - K091107 - K091146 (digest 2) - K091146 - J23116+J37033 (digest 2) - J23116+J37033 - R0010 (digest 2) - R0010
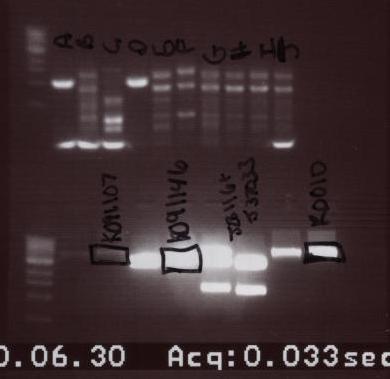
- The bands that were cut are marked with a square around them in the picture above. They were also then purified.
- 3 bands that were cut:
- K091107 - K091146 - R0010
- A second gel was run the same day. The order is provided below:
- Ladder - B1002 - Lux R 2 - C0261+J24679
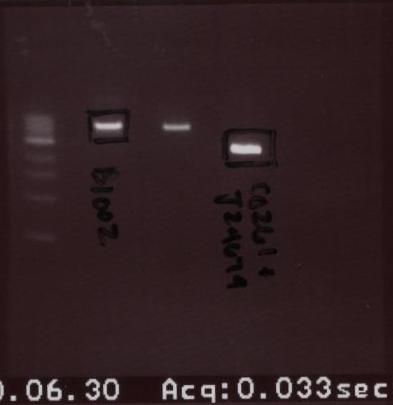
- As you can see, the only 2 bands that were cut were B1002 and C0261+J24679
- Grow for tomorrow:
- K274100 - K112022
July 1
- Flasks minipreped:
- B1006 - K274100 - J23100
- Digests:
- B1006 (EX) - K274100 (XP) - J23100 (SP)
- Sent to the sequencing center:
1) B1002 2) B1006 3) S01414 4) J23100 5) C0261+J24679
- Gel order:
- Ladder - B1006 - K274100 - J23100
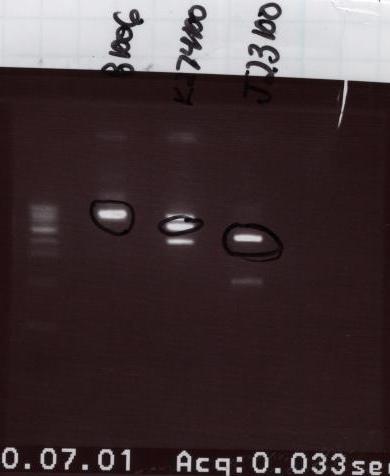
- All the bands appeared where we wanted them to so we cut out all three bands and then purified them.
- Ligations:
| Vector | Inserts |
| 1)J23100 | K274100 |
| 2)J23100 | S01414 |
| 3)R0063 | a)K274100 b)C0261 + J24679 |
| 4)K091146 | a)K274100 b)C0261 + J24679 |
| 5)K091107 | a)K274100 b)C0261 + J24679 |
| 6)R0062 | a)K274100 b)C0261 + J24679 |
| 7)R0010 | K081007 |
- Numbers 3-6 were each matched up with a) K274100 and b) C0261 + J24679. The rest of the vectors were matched with the insert directly to the right in the table.
- Tomorrow:
- Develop primers to figure out if K274100 and K112808 are correct.
July 2
- Colony PCRs run on gel.The order is as follows:
- Ladder - 1A, 1B, 1C - 2A, 2B, 2C - 3A, 3B, 3C - Ladder - 4A, 4B, 4C - 5A, 5B, 5C - 6A, 6B, 6C - 7A, 7B, 7C
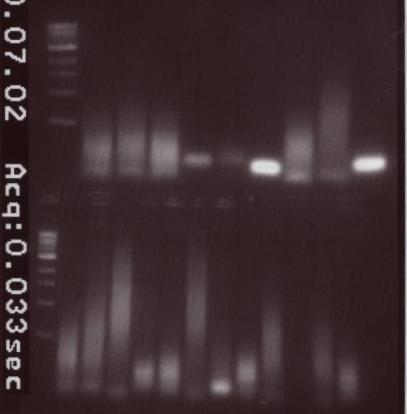
Unfortunately, we accidentally put ladder in all the Colony products. The picture is provided above. We could not use this information in any way other than to learn from our mistake.
 "
"