Team:Davidson-MissouriW/MeasuringExpression
From 2010.igem.org
Theshu0012 (Talk | contribs) |
|||
| (59 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template_Wiki}} | {{Template_Wiki}} | ||
<html> | <html> | ||
| - | + | <h2> iGEM Davidson – MWSU 2010: Measuring Gene Expression </h2> | |
| - | + | <h3> </h3> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | <h3> | + | |
| - | <p> Results from past Davidson and Missouri Western research hinted at the presence of a transcriptional terminator within the TetA gene. Any gene located downstream of the 3’ end of the TetA gene did not appear to be expressed. This gene codes for an effluent pump that physically removes the antibody tetracycline from the cell. | + | <p> The system we originally designed to address the knapsack problem had several vital components. The TetA and RFP genes, along with their corresponding gene products, served as our reporters to see if and how a cell solved the knapsack problem. However, we recorded several observations regarding gene expression as we progressed. Characterizing and attempting to understand these foundational problems became one of the new focuses of this team. |
| - | + | <p> Results from past Davidson and Missouri Western research hinted at the presence of a transcriptional terminator within the TetA gene. Any gene located downstream of the 3’ end of the TetA gene did not appear to be expressed. This gene codes for an effluent pump that physically removes the antibody tetracycline from the cell. To test for the presence of this terminator, we created a construct with a fluorescent protein gene downstream of TetA. If TetA really does block transcription, we would expect to see no fluorescence despite induction of the promoter that precedes the construct (pLac+TetA+RFP). | |
| - | <img src="https://static.igem.org/mediawiki/2010/ | + | <center></p><img src="https://static.igem.org/mediawiki/2010/7/70/Normailized.png" ></center><br> |
| - | <p> The data | + | <p> The data suggests that the order of genes (TetA and RFP) is important as pLac+RFP+TetA shows more fluorescence in comparison to pLac+TetA+RFP. The trend is reproducible when induced by IPTG (0.5mM) as well. Both cell types, however, fluoresced less than the positive control pLac+RFP. The data also supports our hypothesis that a transcriptional terminator is present in the TetA gene. </p> |
| - | <img src="https://static.igem.org/mediawiki/2010/ | + | <center><img src="https://static.igem.org/mediawiki/2010/d/de/2ndpic.png"></center><br> |
| - | <p> | + | <p> In further tests, we observed a significant decrease in fluorescence when the cell type pLac+RFP+TetA was grown in tetracycline (50ug/mL) media. The cells continued to live in the tetracycline media with a marginal drop in cell density. However, the cells displaced considerably less fluorescence in this media even when induced with IPTG. This unexpected result suggests that growth media affects cell fluorescence. |
</p> | </p> | ||
| - | + | <center><img src="https://static.igem.org/mediawiki/2010/6/61/Davidson-MissouriW_RFP_in_JM_109_vs._MG_1655_%282%29.png" id=fig3 height=600 width=800 > | |
| + | </center><br> | ||
| + | <p>MG 1655 E. coli cells have a genotype that closely resembles naturally occurring E. coli. We did a direct comparison of RFP gene expression in MG 1655 cells vs. JM 109. The data we collected is shown in the graph above.</p> | ||
| + | <br> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2010/7/71/Davidson-MissouriW_IPTG_Induction_in_JM_109_vs._MG_1655_%282%29.png" id=fig4 height=600 width=800 > | ||
| + | </center><br> | ||
| + | <p>We also wanted to see if the MG 1655 cells were more prone to induction by IPTG. The data for this experiment is shown in the graph above.</p> | ||
| + | <br> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2010/c/ca/Davidson-MissouriW_Unexplained_Variation_in_Copy_Number_of_RFP_Plasmids_%282%29.png" id=fig5 height=600 width=800 > | ||
| + | </center><br> | ||
| + | <p>While working with two pLac-RBS-RFP clones, we noticed that one of the clones seemed to have a much higher copy number.In order to document the copy number we performed cell density tests and extracted the DNA and determined relative DNA per cell for each clone. The results from this experiment can be viewed above. We do not have an explanation for this variation in copy number.</p> | ||
| + | <br> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2010/2/2b/Davidson-MissouriW_Use_of_LacI_Repressor_to_Achieve_a_Range_of_Induction_%282%29.png" id=fig6 height=600 width=800 > | ||
| + | </center><br> | ||
| + | <p>In order to achieve a range of induction of RFP expression we attached a LacI repressor to the front of a pLac-RBS-RFP-RBS construct. We then proceeded to induce with combinations of IPTG and Ara. Placing the LacI repressor onto to gene affected the level of RFP expression. Results can be seen above.</p> | ||
| + | <br> | ||
| + | |||
| + | <p>During production of a commonly used expression casette, pLac-RBS-RFP, we noticed some colonies that produced a brighter red fluorescence than others. We picked a pink colony and a red colony and sequenced the inserts. We found that the DNA sequences were identical. We traced the difference in phenotype to the inserts by doing experiments that involved switching of the inserts between the two vectors. This led us to suspect that an epigenetic mechanism was involved in this process. The mechanism we proposed was DNA methylation. DNA methylation involves the addition of a methyl group to a nitrogenous base of the DNA. This modification of the DNA can be inherited through subsequent cell divisions and is known to regulate gene expression in a variety of organisms. Adenine or cytosine methylation (Dam or Dcm, respectively) is known to occur in many bacteria, including E. coli. Methylation could affect gene expression by making the DNA specific sequence harder to recognize by the polymerase; thus, decreasing gene expression. Methylation might also increase gene expression by having the opposite effect where the DNA binding site is more easily recognized by the polymerase, though this type is rare. We sought to determine if DNA methylation was actually playing a role in the gene expression of these E. coli cells. Our first experiment used three different restriction endonucleases that were sensitive to DNA methylation to see if any of the sites that are recognized by the enzyme are methylated. This experiment was not able to determine if any DNA in our gene was methylated. A future experiment that will help us in the determination of whether this gene is methylated is methylation-specific PCR (MSP).</p> | ||
| + | <br> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2010/8/8c/Davidson_MWSU_Pink_and_red_clones_vector_switch_for_iGEM.png" id=fig7 height=600 width=800 > | ||
| + | </center><br> | ||
| + | |||
| + | <center><img src="https://static.igem.org/mediawiki/2010/0/08/Davidson-MWSU_vector_switch_on_uv.png" id=fig14 height=600 width=800 > | ||
| + | </center><br> | ||
| + | <p>Top Left = Red clone insert ligated to Red clone vector, Top Right = Red clone insert ligated to Pink clone vector, Bottom Left = Pink clone insert ligated to Red clone vector, Bottom Right = Pink clone insert ligated to Pink clone vector | ||
| + | <p>In another effort to understand why the pLac-RBS-RFP construct was producing one set of red clones and one set of pink clones we designed an experiment to see whether the degree of red fluorescent protein was a function of the vector or the insert. In this experiment we performed a EcoRI/PstI digest on each clone and purified both the vector and insert for each. Then we put each of the inserts back into their original vectors as well as placing the inserts into the opposite vectors. The diagram illustrates the crosses. See pictures of the results of this experiment above. <b>Fluorimeter data confirmed that the intensity of fluorescence in terms of Red or Pick followed the insert, not the vector.</b></p> | ||
| + | <br> | ||
| + | <p></p> | ||
| + | <br> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2010/8/89/Davidson-MWSU_diagram_of_a_fried_egg.png" id="fig10" height=600 width=800 > | ||
| + | </center><br> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2010/2/29/Davidson-MWSU_fried_egg_passage_pic1.png" id="fig11" height=600 width=800 > | ||
| + | </center><br> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2010/9/99/Davidson-MWSU_fried_egg_passage_pic2.png" id="fig12" height=600 width=800 > | ||
| + | </center><br> | ||
| + | <p>As we were doing experiments with a pLac-RBS-RFP construct (part number I715039) we noticed some color variation between clones that were supposed to be identical. In an effort to trace the differences in fluorescence we spotted 2 ml of the two clones. These clones produced ring-like structures resembling a fried egg with a center, a middle, and an outer edge all of different colors. Sections from these colonies were picked, grown up overnight, and spotted again. Each of the new colonies showed similar results. <b>These results suggest that variation in gene expression occurs in clonal populations of bacteria and that these variations are not genetically determined.</b></p> | ||
| + | <br> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2010/e/e7/Davidson-MissouriwesternWaterbroth.png" id="fig9" height=600 width=800 > | ||
| + | </center><br> | ||
| + | <br> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2010/0/07/DavidsonMissouriwesternFlourimeterData.png" id="fig15" height=600 width=800 ></center> | ||
| + | <br> | ||
<center><a href="#main_wrapper">Top<a/></center> | <center><a href="#main_wrapper">Top<a/></center> | ||
| - | |||
| - | |||
<script type="text/javascript"> | <script type="text/javascript"> | ||
| Line 29: | Line 64: | ||
document.getElementById('news').style.height = len + 'px'; | document.getElementById('news').style.height = len + 'px'; | ||
} | } | ||
| - | function | + | |
| - | + | ||
| - | + | function show(id1){ | |
| - | el.style. | + | el=document.getElementById(id1); |
| - | + | if(el.style.display == 'none'){ | |
| + | el.style.display = 'block'; | ||
| + | } | ||
| + | else | ||
| + | el.style.display='none'; | ||
} | } | ||
| + | |||
</script> | </script> | ||
</body></html> | </body></html> | ||
Latest revision as of 01:40, 28 October 2010
iGEM Davidson – MWSU 2010: Measuring Gene Expression
The system we originally designed to address the knapsack problem had several vital components. The TetA and RFP genes, along with their corresponding gene products, served as our reporters to see if and how a cell solved the knapsack problem. However, we recorded several observations regarding gene expression as we progressed. Characterizing and attempting to understand these foundational problems became one of the new focuses of this team.
Results from past Davidson and Missouri Western research hinted at the presence of a transcriptional terminator within the TetA gene. Any gene located downstream of the 3’ end of the TetA gene did not appear to be expressed. This gene codes for an effluent pump that physically removes the antibody tetracycline from the cell. To test for the presence of this terminator, we created a construct with a fluorescent protein gene downstream of TetA. If TetA really does block transcription, we would expect to see no fluorescence despite induction of the promoter that precedes the construct (pLac+TetA+RFP).

The data suggests that the order of genes (TetA and RFP) is important as pLac+RFP+TetA shows more fluorescence in comparison to pLac+TetA+RFP. The trend is reproducible when induced by IPTG (0.5mM) as well. Both cell types, however, fluoresced less than the positive control pLac+RFP. The data also supports our hypothesis that a transcriptional terminator is present in the TetA gene.

In further tests, we observed a significant decrease in fluorescence when the cell type pLac+RFP+TetA was grown in tetracycline (50ug/mL) media. The cells continued to live in the tetracycline media with a marginal drop in cell density. However, the cells displaced considerably less fluorescence in this media even when induced with IPTG. This unexpected result suggests that growth media affects cell fluorescence.

MG 1655 E. coli cells have a genotype that closely resembles naturally occurring E. coli. We did a direct comparison of RFP gene expression in MG 1655 cells vs. JM 109. The data we collected is shown in the graph above.

We also wanted to see if the MG 1655 cells were more prone to induction by IPTG. The data for this experiment is shown in the graph above.

While working with two pLac-RBS-RFP clones, we noticed that one of the clones seemed to have a much higher copy number.In order to document the copy number we performed cell density tests and extracted the DNA and determined relative DNA per cell for each clone. The results from this experiment can be viewed above. We do not have an explanation for this variation in copy number.
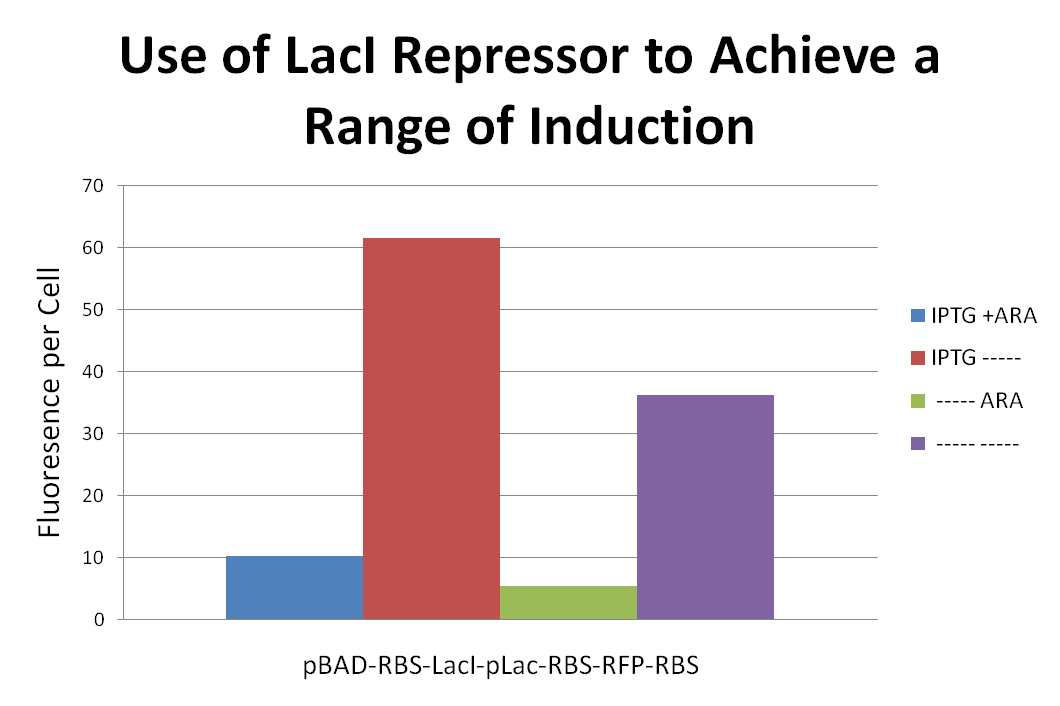
In order to achieve a range of induction of RFP expression we attached a LacI repressor to the front of a pLac-RBS-RFP-RBS construct. We then proceeded to induce with combinations of IPTG and Ara. Placing the LacI repressor onto to gene affected the level of RFP expression. Results can be seen above.
During production of a commonly used expression casette, pLac-RBS-RFP, we noticed some colonies that produced a brighter red fluorescence than others. We picked a pink colony and a red colony and sequenced the inserts. We found that the DNA sequences were identical. We traced the difference in phenotype to the inserts by doing experiments that involved switching of the inserts between the two vectors. This led us to suspect that an epigenetic mechanism was involved in this process. The mechanism we proposed was DNA methylation. DNA methylation involves the addition of a methyl group to a nitrogenous base of the DNA. This modification of the DNA can be inherited through subsequent cell divisions and is known to regulate gene expression in a variety of organisms. Adenine or cytosine methylation (Dam or Dcm, respectively) is known to occur in many bacteria, including E. coli. Methylation could affect gene expression by making the DNA specific sequence harder to recognize by the polymerase; thus, decreasing gene expression. Methylation might also increase gene expression by having the opposite effect where the DNA binding site is more easily recognized by the polymerase, though this type is rare. We sought to determine if DNA methylation was actually playing a role in the gene expression of these E. coli cells. Our first experiment used three different restriction endonucleases that were sensitive to DNA methylation to see if any of the sites that are recognized by the enzyme are methylated. This experiment was not able to determine if any DNA in our gene was methylated. A future experiment that will help us in the determination of whether this gene is methylated is methylation-specific PCR (MSP).


Top Left = Red clone insert ligated to Red clone vector, Top Right = Red clone insert ligated to Pink clone vector, Bottom Left = Pink clone insert ligated to Red clone vector, Bottom Right = Pink clone insert ligated to Pink clone vector
In another effort to understand why the pLac-RBS-RFP construct was producing one set of red clones and one set of pink clones we designed an experiment to see whether the degree of red fluorescent protein was a function of the vector or the insert. In this experiment we performed a EcoRI/PstI digest on each clone and purified both the vector and insert for each. Then we put each of the inserts back into their original vectors as well as placing the inserts into the opposite vectors. The diagram illustrates the crosses. See pictures of the results of this experiment above. Fluorimeter data confirmed that the intensity of fluorescence in terms of Red or Pick followed the insert, not the vector.



As we were doing experiments with a pLac-RBS-RFP construct (part number I715039) we noticed some color variation between clones that were supposed to be identical. In an effort to trace the differences in fluorescence we spotted 2 ml of the two clones. These clones produced ring-like structures resembling a fried egg with a center, a middle, and an outer edge all of different colors. Sections from these colonies were picked, grown up overnight, and spotted again. Each of the new colonies showed similar results. These results suggest that variation in gene expression occurs in clonal populations of bacteria and that these variations are not genetically determined.


 "
"