Team:Stockholm/27 July 2010
From 2010.igem.org
(New page: {{Stockholm/Top2}} Image:Net image a jM9yggVoo68J high res.jpg) |
m |
||
| (12 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Stockholm/Top2}} | {{Stockholm/Top2}} | ||
| - | [[Image:Net image a jM9yggVoo68J high res.jpg]] | + | |
| + | == Mimmi == | ||
| + | |||
| + | |||
| + | === over-expression in pEX === | ||
| + | ::::- '''remaking the comassie-gel''' | ||
| + | |||
| + | |||
| + | |||
| + | {| border="1" cellspacing="0" | ||
| + | |- | ||
| + | ! Mix | ||
| + | | v | ||
| + | |- | ||
| + | | LB | ||
| + | | 4 ml | ||
| + | |- | ||
| + | | glycose | ||
| + | | 40 µl (1%) | ||
| + | |- | ||
| + | | Amp | ||
| + | | 8 µl (0.5%) | ||
| + | |- | ||
| + | | old culture | ||
| + | | 40 µl | ||
| + | |} | ||
| + | |||
| + | |||
| + | *Grow in 37C, ~200 rpm ~2h until OD = 0.6 | ||
| + | |||
| + | *Add IPTG (1 µl/ml,1M)in one of the two cultures, the other one is used as a control | ||
| + | |||
| + | *Take sample at 0h, 1h, 2h, 3h | ||
| + | **pipette 500µl into a 1.5ml tube | ||
| + | **spinn down the sells and remove LB | ||
| + | **resuspend in 50µl loading dye | ||
| + | **freeze | ||
| + | |||
| + | |||
| + | |||
| + | === Site-Directed Mutagenesis === | ||
| + | |||
| + | ==== Primers ==== | ||
| + | :(MW/10)/(OD*33) = dilution factor | ||
| + | ::1000/df = volyme to get 100µM primer | ||
| + | :::100µM = (MW/10) ng/µl | ||
| + | ::::(MW/10)/125 = df to get 125 ng/µl | ||
| + | |||
| + | ::{| border="1" cellspacing="0" | ||
| + | |- | ||
| + | ! primer | ||
| + | ! V H2O (µl) | ||
| + | ! V H2O to get 125 ng/µl (µl) | ||
| + | |- | ||
| + | | MITF_site1_F | ||
| + | | 463.88 | ||
| + | | 3µl + 15.4µl | ||
| + | |- | ||
| + | | MITF_site1_R | ||
| + | | 64.77 | ||
| + | | 3µl + 15.3µl | ||
| + | |- | ||
| + | | MITF_site2_F | ||
| + | | 144.85 | ||
| + | | 3µl + 21µl | ||
| + | |- | ||
| + | | MITF_site2_R | ||
| + | | 141.77 | ||
| + | | 3µl + 21.6µl | ||
| + | |- | ||
| + | | | ||
| + | ! concentration | ||
| + | | | ||
| + | |- | ||
| + | | yCCS_F | ||
| + | | 1085.7 ng/µl | ||
| + | | 3µl + 23µl | ||
| + | |- | ||
| + | | yCCS_R | ||
| + | | 996.5 ng/µl | ||
| + | | 3µl + 20.9µl | ||
| + | |} | ||
| + | |||
| + | ==== reaction ==== | ||
| + | |||
| + | {| border="1" cellspacing="0" | ||
| + | |- | ||
| + | ! Mix | ||
| + | | width="50" | (µl) | ||
| + | | width="50" |X2 | ||
| + | |- | ||
| + | | H2O | ||
| + | | 40 | ||
| + | | 80 | ||
| + | |- | ||
| + | | dNTPs | ||
| + | | 1 | ||
| + | | 2 | ||
| + | |- | ||
| + | | F primer | ||
| + | | 1 | ||
| + | | 2 | ||
| + | |- | ||
| + | | R primer | ||
| + | | 1 | ||
| + | | 2 | ||
| + | |- | ||
| + | | DNA | ||
| + | | 1 | ||
| + | | 2*1 | ||
| + | |- | ||
| + | | Pfu X10 buffer | ||
| + | | 5 | ||
| + | | 10 | ||
| + | |- | ||
| + | | Pfu turbo pol | ||
| + | | 1 | ||
| + | | 2 | ||
| + | |} | ||
| + | :::::::{| border="0" width="50%" | | ||
| + | ! align="left" | MITF | ||
| + | ! align="left" | yCCS | ||
| + | ! conditions | ||
| + | |- | ||
| + | |MITF_Site1_F | ||
| + | |yCCS_F | ||
| + | !align="left" width="1%"|time | ||
| + | !align="left" width="1%"|°C | ||
| + | |- | ||
| + | |MITF_Site2_R | ||
| + | |yCCS_R | ||
| + | |30s | ||
| + | |95 | ||
| + | |- | ||
| + | |pRc/CMV | ||
| + | |pSB1C3 | ||
| + | |30s | ||
| + | |95 | ||
| + | |- | ||
| + | |~6.7kb | ||
| + | |~3,5kb | ||
| + | |30s | ||
| + | |55 | ||
| + | |- | ||
| + | | | ||
| + | | | ||
| + | |7m | ||
| + | |68 | ||
| + | |- | ||
| + | | | ||
| + | | | ||
| + | |oo | ||
| + | |4 | ||
| + | |} | ||
| + | |||
| + | |||
| + | ::*Make sure sample is ≤ 37°C | ||
| + | ::*Add 1µl Dpn1 (to 50µl product) and incubate in 37°C ON | ||
| + | |||
| + | |||
| + | |||
| + | ===Transformation=== | ||
| + | |||
| + | ::<u>mix</u> | ||
| + | ::Top 10 competent cells 100µl | ||
| + | ::pRc/CMV.MITF_M 1µl | ||
| + | |||
| + | |||
| + | *Hold on ice 30min | ||
| + | *Heat shock 42°C, 55sec | ||
| + | *Cool down 1min (on ice) | ||
| + | *Add 900µl LB | ||
| + | *Incubate 37°C, 250rpm, 1h (forgot rpm) | ||
| + | *Spinn down cells, 13000rpm, 15sec | ||
| + | *Remove 900µl LB | ||
| + | *Plate the 100µl on a Amp-plate | ||
| + | *Grow ON at 37°C | ||
| + | |||
| + | ==Hassan== | ||
| + | |||
| + | [[Image:Net image a jM9yggVoo68J high res.jpg|500px|thumb|left|Jensen et al. Nucleic Acids Res. 2009, 37(Database issue):D412-6]] | ||
| + | |||
| + | [[Image:Net image a V5uyW3lEbFAM.jpg|400px|thumb|left|Jensen et al. Nucleic Acids Res. 2009, 37(Database issue):D412-6]] | ||
| + | |||
| + | ---- | ||
| + | |||
| + | [http://string.embl.de/version_8_3/newstring_cgi/show_network_section.pl?identifiers=9606.ENSP00000331746%250D9606.ENSP00000295600%250D9606.ENSP00000364016%250D9606.ENSP00000371175%250D9606.ENSP00000269280%250D9606.ENSP00000363700%250D9606.ENSP00000360157%250D9606.ENSP00000373571%250D9606.ENSP00000291700&channel1=off&channel2=off&channel3=off&channel4=on&channel5=on&channel6=on&channel7=on&interactive=yes&network_flavor=actions&targetmode=proteins] | ||
| + | |||
| + | [http://string.embl.de/version_8_3/newstring_cgi/show_network_section.pl?identifiers=9606.ENSP00000331746%250D9606.ENSP00000295600%250D9606.ENSP00000226877%250D9606.ENSP00000371175%250D9606.ENSP00000269280%250D9606.ENSP00000363700%250D9606.ENSP00000360157%250D9606.ENSP00000373571%250D9606.ENSP00000291700&channel1=off&channel2=off&channel3=off&channel4=on&channel5=on&channel6=on&channel7=on&additional_network_nodes=5&interactive=yes&network_flavor=actions&targetmode=proteins] | ||
| + | |||
| + | |||
| + | ---- | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ---- | ||
| + | |||
| + | ---- | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ---- | ||
| + | |||
| + | ---- | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ---- | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ---- | ||
| + | |||
| + | |||
| + | ---- | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ---- | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ---- | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ---- | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ---- | ||
| + | |||
| + | ==Nina== | ||
| + | |||
| + | ==Colony PCR on IgG protease in shipping vector== | ||
| + | |||
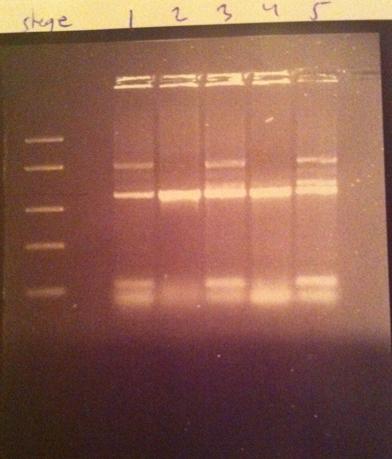
| + | I made a colony PCR on the IgG protease that I have inserted into the iGEM shipping vector to verify that the gene is inserted in a correct position. Therefore I used one of the gene's primers and one of the vector's verification primers. The colony numbers are: 1, 2, 3, 4 and 5. | ||
| + | |||
| + | PCR reaction mix: | ||
| + | |||
| + | * 1 µl Morten's polymerase PjuX7 | ||
| + | |||
| + | * 1 µl 10 mM dNTPs | ||
| + | |||
| + | * 3 µl 5 µM forward primer (VF2) | ||
| + | |||
| + | * 3 µl 5 µM revers primer (gene's primer) | ||
| + | |||
| + | * 10 µl buffer 5X | ||
| + | |||
| + | * 1 µl MgCl2 50mM | ||
| + | |||
| + | * 30 µl H2O | ||
| + | |||
| + | * DNA template was one colony | ||
| + | |||
| + | PCR program: | ||
| + | |||
| + | 98°C - 2 min | ||
| + | |||
| + | 31 cycles of: | ||
| + | |||
| + | * 98°C - 10 sec | ||
| + | |||
| + | * 55°C - 15 sec | ||
| + | |||
| + | * 72°C - 1.5 min | ||
| + | |||
| + | 72°C - 5 min | ||
| + | |||
| + | 4°C - ∞ | ||
| + | |||
| + | [[Image:Igg-shipping.jpg|250px]] | ||
| + | |||
| + | DNA Ladder: FastRuler™ Middle Range, ready-to-use, 100-5000 bp Fermentas | ||
| + | |||
| + | Colony number 2 looks good on the gel. This one will be inoculated in LB in order to become minipreped to be shipped to iGEM hq. | ||
| + | |||
| + | ---- | ||
| + | |||
| + | ==Mini prep on IgG protease and CPP== | ||
| + | |||
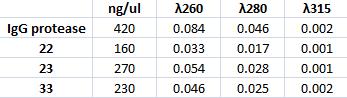
| + | I prepare a miniprep on the inoculated IgG protease with colony number 2. In addition I miniprep three inoculated samples with vector carrying CPP from colony number 22, 23 and 33. | ||
| + | |||
| + | The method is carried out according to the procedure in protocols. | ||
| + | |||
| + | Measuring concentration with spectrophotometer: | ||
| + | |||
| + | [[Image:Spek.jpg]] | ||
| + | |||
| + | ---- | ||
| + | |||
| + | ==Sequencing CPP TAT N version== | ||
| + | |||
| + | I send three samples of CPP TAT N version for sequencing. Colony numbers are: 22, 23 and 33. | ||
| + | |||
| + | * 15 ul vector | ||
| + | |||
| + | * 1.5 ul 10uM VR2 primer | ||
| + | |||
| + | 22: ASB0045 105 | ||
| + | |||
| + | 23: ASB0045 104 | ||
| + | |||
| + | 33: ASB0045 103 | ||
| + | |||
| + | {{Stockholm/Footer}} | ||
Latest revision as of 10:56, 26 October 2010
Contents |
Mimmi
over-expression in pEX
- - remaking the comassie-gel
| Mix | v |
|---|---|
| LB | 4 ml |
| glycose | 40 µl (1%) |
| Amp | 8 µl (0.5%) |
| old culture | 40 µl |
- Grow in 37C, ~200 rpm ~2h until OD = 0.6
- Add IPTG (1 µl/ml,1M)in one of the two cultures, the other one is used as a control
- Take sample at 0h, 1h, 2h, 3h
- pipette 500µl into a 1.5ml tube
- spinn down the sells and remove LB
- resuspend in 50µl loading dye
- freeze
Site-Directed Mutagenesis
Primers
- (MW/10)/(OD*33) = dilution factor
- 1000/df = volyme to get 100µM primer
- 100µM = (MW/10) ng/µl
- (MW/10)/125 = df to get 125 ng/µl
- 100µM = (MW/10) ng/µl
- 1000/df = volyme to get 100µM primer
primer V H2O (µl) V H2O to get 125 ng/µl (µl) MITF_site1_F 463.88 3µl + 15.4µl MITF_site1_R 64.77 3µl + 15.3µl MITF_site2_F 144.85 3µl + 21µl MITF_site2_R 141.77 3µl + 21.6µl concentration yCCS_F 1085.7 ng/µl 3µl + 23µl yCCS_R 996.5 ng/µl 3µl + 20.9µl
reaction
| Mix | (µl) | X2 |
|---|---|---|
| H2O | 40 | 80 |
| dNTPs | 1 | 2 |
| F primer | 1 | 2 |
| R primer | 1 | 2 |
| DNA | 1 | 2*1 |
| Pfu X10 buffer | 5 | 10 |
| Pfu turbo pol | 1 | 2 |
MITF yCCS conditions MITF_Site1_F yCCS_F time °C MITF_Site2_R yCCS_R 30s 95 pRc/CMV pSB1C3 30s 95 ~6.7kb ~3,5kb 30s 55 7m 68 oo 4
- Make sure sample is ≤ 37°C
- Add 1µl Dpn1 (to 50µl product) and incubate in 37°C ON
Transformation
- mix
- Top 10 competent cells 100µl
- pRc/CMV.MITF_M 1µl
- Hold on ice 30min
- Heat shock 42°C, 55sec
- Cool down 1min (on ice)
- Add 900µl LB
- Incubate 37°C, 250rpm, 1h (forgot rpm)
- Spinn down cells, 13000rpm, 15sec
- Remove 900µl LB
- Plate the 100µl on a Amp-plate
- Grow ON at 37°C
Hassan
[http://string.embl.de/version_8_3/newstring_cgi/show_network_section.pl?identifiers=9606.ENSP00000331746%250D9606.ENSP00000295600%250D9606.ENSP00000364016%250D9606.ENSP00000371175%250D9606.ENSP00000269280%250D9606.ENSP00000363700%250D9606.ENSP00000360157%250D9606.ENSP00000373571%250D9606.ENSP00000291700&channel1=off&channel2=off&channel3=off&channel4=on&channel5=on&channel6=on&channel7=on&interactive=yes&network_flavor=actions&targetmode=proteins]
[http://string.embl.de/version_8_3/newstring_cgi/show_network_section.pl?identifiers=9606.ENSP00000331746%250D9606.ENSP00000295600%250D9606.ENSP00000226877%250D9606.ENSP00000371175%250D9606.ENSP00000269280%250D9606.ENSP00000363700%250D9606.ENSP00000360157%250D9606.ENSP00000373571%250D9606.ENSP00000291700&channel1=off&channel2=off&channel3=off&channel4=on&channel5=on&channel6=on&channel7=on&additional_network_nodes=5&interactive=yes&network_flavor=actions&targetmode=proteins]
Nina
Colony PCR on IgG protease in shipping vector
I made a colony PCR on the IgG protease that I have inserted into the iGEM shipping vector to verify that the gene is inserted in a correct position. Therefore I used one of the gene's primers and one of the vector's verification primers. The colony numbers are: 1, 2, 3, 4 and 5.
PCR reaction mix:
- 1 µl Morten's polymerase PjuX7
- 1 µl 10 mM dNTPs
- 3 µl 5 µM forward primer (VF2)
- 3 µl 5 µM revers primer (gene's primer)
- 10 µl buffer 5X
- 1 µl MgCl2 50mM
- 30 µl H2O
- DNA template was one colony
PCR program:
98°C - 2 min
31 cycles of:
- 98°C - 10 sec
- 55°C - 15 sec
- 72°C - 1.5 min
72°C - 5 min
4°C - ∞
DNA Ladder: FastRuler™ Middle Range, ready-to-use, 100-5000 bp Fermentas
Colony number 2 looks good on the gel. This one will be inoculated in LB in order to become minipreped to be shipped to iGEM hq.
Mini prep on IgG protease and CPP
I prepare a miniprep on the inoculated IgG protease with colony number 2. In addition I miniprep three inoculated samples with vector carrying CPP from colony number 22, 23 and 33.
The method is carried out according to the procedure in protocols.
Measuring concentration with spectrophotometer:
Sequencing CPP TAT N version
I send three samples of CPP TAT N version for sequencing. Colony numbers are: 22, 23 and 33.
- 15 ul vector
- 1.5 ul 10uM VR2 primer
22: ASB0045 105
23: ASB0045 104
33: ASB0045 103
 |

|
 |

|
 |

|

|

|
 "
"