Team:Cambridge/References/ProjectBioluminescence
From 2010.igem.org
(→Sequences) |
|||
| (33 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{:Team:Cambridge/Templates/ | + | {{:Team:Cambridge/Templates/headerMinimalprototype}} |
| - | {{:Team:Cambridge/ | + | {{:Team:Cambridge/Templates/RefBar}} |
| - | = | + | {{:Team:Cambridge/Templates/headerbar|colour=#96d446|title=Bioluminescence: Introduction}} |
| + | |||
==The Plan== | ==The Plan== | ||
* Characterise the transfer function of PoPs + CHBT --> Light | * Characterise the transfer function of PoPs + CHBT --> Light | ||
| + | * Our Characterisation should look like [http://www.nature.com/nbt/journal/v26/n7/abs/nbt1413.html this]. | ||
| + | ==The Theory== | ||
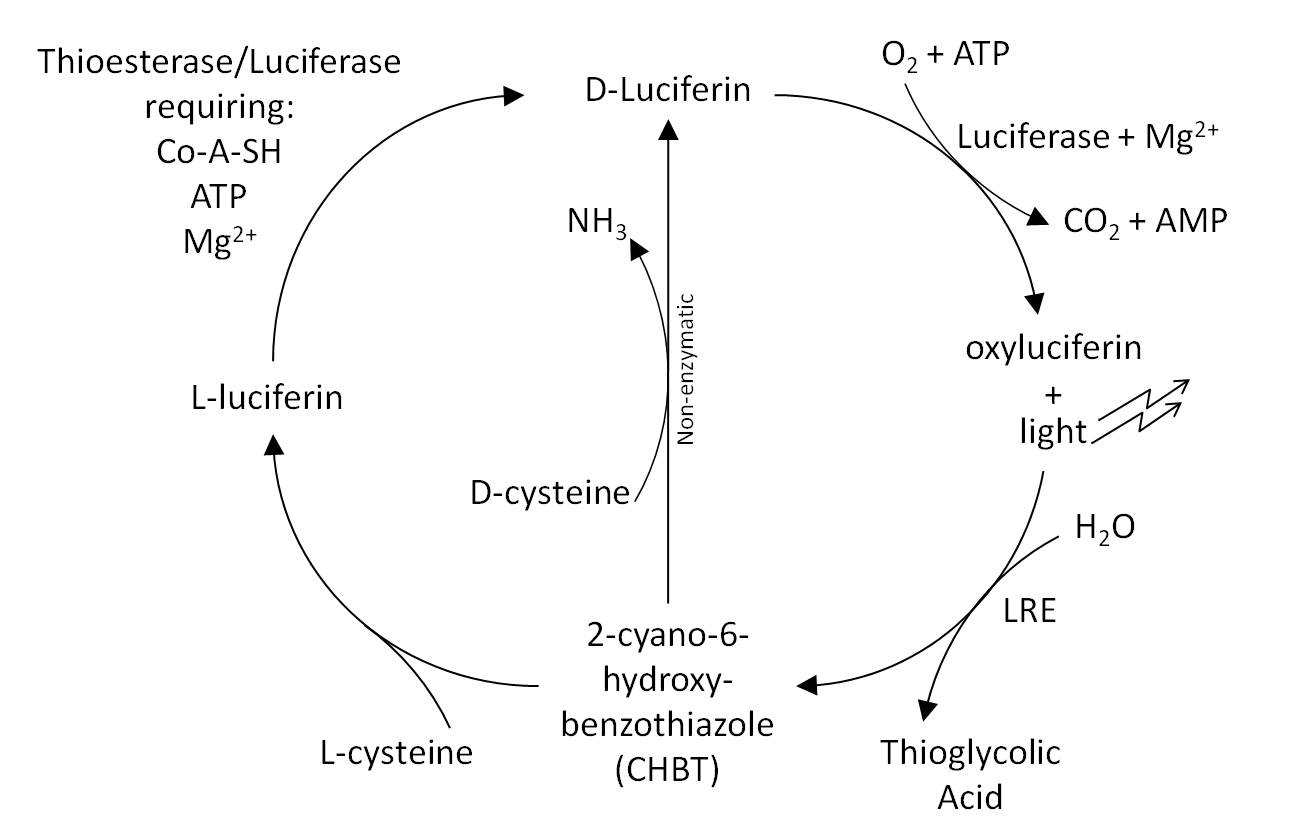
* The Luciferin Cycle: | * The Luciferin Cycle: | ||
[[Image:Cam-luci-cycle.jpg | 600px]] | [[Image:Cam-luci-cycle.jpg | 600px]] | ||
| - | + | ==Amazing mutant== | |
| - | + | paper on cloning the lux operon from Ponyfish Photobacterium leiognathi into E.coli, by the E.coli mutant 43R the luminescence output could be increased dramatically to near native levels. --> find out what 43R does! [http://www3.interscience.wiley.com/cgi-bin/fulltext/120766166/PDFSTART Chan et al. 1991] suspects negative effectors in wild-type E.coli, but not in the mutant or in P.leiognathi, inhibit luminescence. If this is the case, it might be difficult to mimic the mutant phenotype with genes imported on a plasmid. | |
| - | + | [http://www.springerlink.com/content/vlxb2c59xut1htd8/fulltext.pdf Ulitzur et al. 1997] describes phenotypes like 43R being generated by deletions in the H-NS gene, fancy that. Apparently H-NS acts as a pleiotropic transcriptional repressor, with particularly strong effects on a certain set of promoters. Mutants have a reduced expression of a flagellar protein and are non-motile, but fully viable. In fact they appear to grow faster. However, H-NS has been implicated in E.coli thermostability. | |
| - | [http://www. | + | |
| - | + | ||
| - | {{:Team:Cambridge/Templates/ | + | ==ADLA== |
| + | The Abrupt Decline in Luciferase Activity is a phenomenon described for bacterial luciferase in E.coli in stationary phase of growth (see [http://www.springerlink.com/content/w73k840k27866462/fulltext.pdf Koga et al. 2005]) The paper identifies two mutants, HupA and an N-terminal deletion of H-NS for which ADLA does not occur. hns205, the mutation necessary for quiescence is a C-terminal deletion. We should find out whether the phenotypes are the same, which would be sweet if we ended up doing quiescence as well. | ||
| + | <html> | ||
| + | </div> | ||
| + | </html> | ||
| + | {{:Team:Cambridge/Templates/footerMinimal}} | ||
Latest revision as of 12:16, 7 October 2010

The Plan
- Characterise the transfer function of PoPs + CHBT --> Light
- Our Characterisation should look like [http://www.nature.com/nbt/journal/v26/n7/abs/nbt1413.html this].
The Theory
- The Luciferin Cycle:
Amazing mutant
paper on cloning the lux operon from Ponyfish Photobacterium leiognathi into E.coli, by the E.coli mutant 43R the luminescence output could be increased dramatically to near native levels. --> find out what 43R does! [http://www3.interscience.wiley.com/cgi-bin/fulltext/120766166/PDFSTART Chan et al. 1991] suspects negative effectors in wild-type E.coli, but not in the mutant or in P.leiognathi, inhibit luminescence. If this is the case, it might be difficult to mimic the mutant phenotype with genes imported on a plasmid.
[http://www.springerlink.com/content/vlxb2c59xut1htd8/fulltext.pdf Ulitzur et al. 1997] describes phenotypes like 43R being generated by deletions in the H-NS gene, fancy that. Apparently H-NS acts as a pleiotropic transcriptional repressor, with particularly strong effects on a certain set of promoters. Mutants have a reduced expression of a flagellar protein and are non-motile, but fully viable. In fact they appear to grow faster. However, H-NS has been implicated in E.coli thermostability.
ADLA
The Abrupt Decline in Luciferase Activity is a phenomenon described for bacterial luciferase in E.coli in stationary phase of growth (see [http://www.springerlink.com/content/w73k840k27866462/fulltext.pdf Koga et al. 2005]) The paper identifies two mutants, HupA and an N-terminal deletion of H-NS for which ADLA does not occur. hns205, the mutation necessary for quiescence is a C-terminal deletion. We should find out whether the phenotypes are the same, which would be sweet if we ended up doing quiescence as well.
 "
"