Team:KAIST-Korea/Project/Introduction
From 2010.igem.org
(→Fusion Antibody Receptor Making) |
|||
| (77 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | |||
{{:Team:KAIST/header}} | {{:Team:KAIST/header}} | ||
| - | + | <table width="100%" border="0" cellpadding="20px"> | |
| - | + | ||
| - | <table width="100%" border="0"> | + | |
| - | + | ||
<tr> | <tr> | ||
| - | |||
<td valign="top" width="75%"> | <td valign="top" width="75%"> | ||
| - | == Motivation | + | <span style=font-size:30px><font face="Maiandra GD"><b>Motivation </b></font></span> |
| - | + | ||
<br> | <br> | ||
| - | [[Image:Estimated TB incidence rates.jpg|center]] | + | ---- |
| + | <br> | ||
| + | We are suffering from various kinds of diseases. Three most prevalent ones would be malaria, HIV and tuberculosis. Malaria is transmitted from human to human by mosquitoes. Each year, malaria causes nearly one million deaths, and the majority of casualties occur in Africa. Furthermore, disease is high-risk in children, pregnant women, travelers, refugees, displaced persons, and laborers entering endemic areas. In the case of HIV, there are 33 million infected people in the world and 95 percent of them are living in developing countries. Last but not least, there is tuberculosis, which is caused by mycobacterium tuberculosis, affecting lungs most. Until now, more than 2 billion people are infected with tuberculosis bacilli and 90 percent of them live in developing countries. As we can see, all of these diseases are especially problematic in developing country. | ||
| + | <br> | ||
| + | [[Image:Estimated TB incidence rates.jpg|center|600px]] | ||
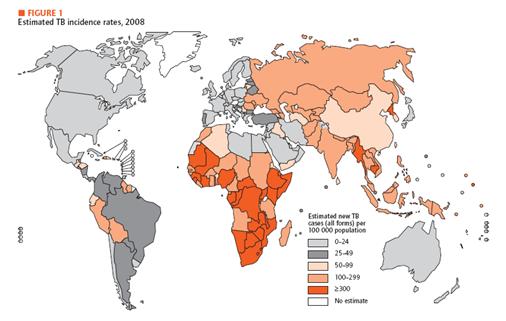
<span style =font-size:15px><b><center>Fig 1. Estimated TB incidence rates, 2008 </center></b></span> | <span style =font-size:15px><b><center>Fig 1. Estimated TB incidence rates, 2008 </center></b></span> | ||
<br> | <br> | ||
| - | In developing country, | + | In developing country, economic is unstable, and development in medical technology is not a priority. Unfortunately, basic medical examination like blood tests, biopsy and others need certain foundation of medical technology. In addition, some areas in developing countries do not offer electricity or access to high-tech equipments. Therefore, we need new and affordable diagnosis system especially for the diseases mentioned above. For our system, we choose TB as a target. In recent decades, diagnosis and treatment of TB have been developed. Diagnostic methods include Tuberculin Skin Test, X-ray Test for its activation and examination of the sputum. In addition, people take a blood test that involves ESR, white blood cell, CRP status checking. If necessary, CT scan and endoscopy can be performed. Most of these tests, however, require high technology and proper machines. Therefore, in developing countries, these tests are not really options. As an alternative, we introduce DiscoverY. |
| - | <br> | + | <br><br> |
| - | [[Image:suffering.jpg| | + | [[Image:suffering.jpg|800px|center]] |
| - | <span style =font-size:15px><b><center>Fig 2. People suffering from malaria and TB | + | <span style =font-size:15px><b><center>Fig 2. People suffering from malaria and TB Fig 3. Researching disease</center></b></span> |
<br> | <br> | ||
| - | | + | We suggest a system using antigen-antibody reaction. Actually, antibody cannot release perceivable signals by antigen-antibody reaction alone, so we thought of fusion antibody-receptor as the solution. Fusion antibody-receptor is a receptor which is fused with other functional protein, antibody. In this way, we can get result by gene expression which would generate GFP expression after our system is triggered by target antigens. As the result, we may develop universal sensor by changing only target antigen’s antibody in fusion antibody-receptor. It will provide relatively simple and direct way of diagnosis. |
| - | <br> | + | <br><br> |
[[Image:antibody.jpg|center]] | [[Image:antibody.jpg|center]] | ||
<span style =font-size:15px><b><center>Fig 4. Antibody</center></b></span> | <span style =font-size:15px><b><center>Fig 4. Antibody</center></b></span> | ||
<br> | <br> | ||
<br> | <br> | ||
| - | + | <br> | |
| - | == Whole Pathway | + | <br> |
| - | + | <span style=font-size:30px><font face="Maiandra GD"><b>Whole Pathway </b></font></span> | |
| + | <br> | ||
| + | ---- | ||
| + | <br> | ||
<html> | <html> | ||
| - | <center><embed src="https://static.igem.org/mediawiki/2010/ | + | <center><embed src="https://static.igem.org/mediawiki/2010/8/89/Working_final.swf" width=550 height=400></embed></center> |
</html> | </html> | ||
| + | <br> | ||
| + | <table align=center> | ||
| + | <tr><td> | ||
| + | <span style=font-size:15px> <b> DiscoverY pathway </b> </span> | ||
| + | </td></tr> | ||
| + | <tr><td> | ||
| + | 1. 'DiscoverY' is spread on the sample.</td></tr> | ||
| + | <tr><td> | ||
| + | 2. 'DiscoverY’s antibody combines with the antigen on the ''Mycobacterium tuberculosis''.</td></tr> | ||
| + | <tr><td> | ||
| + | 3. The tyrosine kinase receptor FGFR binds to the cytokine receptor integral to membrane.</td></tr> | ||
| + | <tr><td> | ||
| + | 4. FGFR autophosphorylation activates transcription factors called STAT1.</td></tr> | ||
| + | <tr><td> | ||
| + | 5. The activated STAT1 dissociates from the receptor.</td></tr> | ||
| + | <tr><td> | ||
| + | 6. The activated STAT1 forms dimers.</td></tr> | ||
| + | <tr><td> | ||
| + | 7. The STAT1 dimer goes into nucleus and regulates GFP gene that is attached to GAS elements of promoter.</td></tr> | ||
| + | <tr><td> | ||
| + | 8. 'DiscoverY' expresses GFP.</td></tr> | ||
| + | </table> | ||
<br><br> | <br><br> | ||
| - | + | <br><br> | |
| - | == Antibodies for Detecting ''Mycobacterium tuberculosis'' | + | <span style=font-size:30px><font face="Maiandra GD"><b>Antibodies for Detecting<br><br> ''Mycobacterium tuberculosis'' </b></font></span> |
| - | The major problem of Tuberculosis | + | <br> |
| + | ---- | ||
| + | <br> | ||
| + | The major problem of Tuberculosis includes the prevalence of multi-drug-resistant strains and co-infection with human immunodeficiency virus. We found a report claiming that <b>MPT51</b> is over-expressed on the tuberculosis cell membrane, so we can use MPT51 as a core antigen to detect tuberculosis. Also, other normal human cell in our body does not have MPT51 protein on a cell. In conclusion, existence of MPT51 on a cell tells if subject has tuberculosis or not. Therefore, we will make fusion antibody that can cognize MPT51 as an antigen, and put it into yeast. | ||
<br> | <br> | ||
<br> | <br> | ||
| Line 45: | Line 71: | ||
<br> | <br> | ||
<br> | <br> | ||
| - | + | <br> | |
| - | == Making Biosensor – Yeast Detector | + | <br> |
| + | <span style=font-size:30px><font face="Maiandra GD"><b>Making Biosensor – Yeast Detector </b></font></span> | ||
| + | <br> | ||
| + | ---- | ||
| + | <br> | ||
=== Biosensor === | === Biosensor === | ||
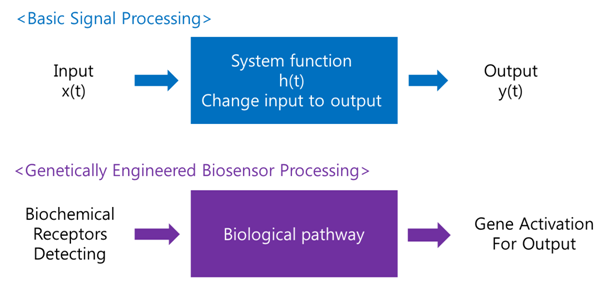
A '''biosensor''' is a device for the detection of an analyst that combines a biological component with a physicochemical detector component.[1] In order to make biosensor with genetically engineered organism, we should consider the following basic sensing process. | A '''biosensor''' is a device for the detection of an analyst that combines a biological component with a physicochemical detector component.[1] In order to make biosensor with genetically engineered organism, we should consider the following basic sensing process. | ||
<center><u>Receive input -> Process -> Extract output</u></center> | <center><u>Receive input -> Process -> Extract output</u></center> | ||
| - | The ‘Receive input’ part can be alternated by represented receptor, the ‘Process’ part can be signaling pathway which changes input to output, and the ‘Extract output’ part can be something easily detectable by human. Nowadays, pigments or fluorescence proteins are usually used for output, and in order to express these substances, | + | The ‘Receive input’ part can be alternated by represented receptor, the ‘Process’ part can be signaling pathway which changes input to output, and the ‘Extract output’ part can be something easily detectable by human. Nowadays, pigments or fluorescence proteins are usually used for output, and, in order to express these substances, we make genetically modified organism. Thus, all engineered biosensors with organisms should be modeled on (consist of) '''1) biochemical receptors''' which can activate gene expression, '''2) biological pathway''', and '''3) gene activation''' for output. |
<br><br> | <br><br> | ||
[[Image:Int4.png|Biosensor Processing|center]] | [[Image:Int4.png|Biosensor Processing|center]] | ||
<br><br><br> | <br><br><br> | ||
=== Organism Decision : Why Yeast? === | === Organism Decision : Why Yeast? === | ||
| - | Why we use ‘Yeast’ instead of ‘E. coli’? | + | Why we use ‘Yeast’ instead of ‘E. coli’? E. coli control is easier than Yeast control. E. coli has faster cell cycle than Yeast. Moreover, there are many E. coli templates. But, although E. coli has many profits, we choose Yeast. Why? The following table is E. coli properties.[2] |
| - | E. coli control is easier than Yeast control. E. coli has faster cell cycle than Yeast. Moreover, there are many E. coli templates. But, although E. coli has many profits, we choose Yeast. Why? The following table is E. coli properties.[2] | + | |
<br><br> | <br><br> | ||
[[Image:E.coli general statistics.jpg|500px|center]] | [[Image:E.coli general statistics.jpg|500px|center]] | ||
| + | |||
| + | <span style=font-size:15px><center><b>Fig 6. E.coli statistics</b></center></span> | ||
<br> | <br> | ||
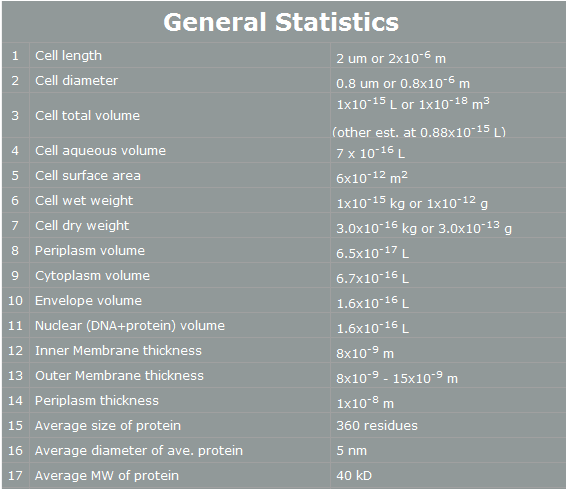
| - | According to the table, we can | + | According to the table, we can easily find reasons why E. coli cannot support human pathway well.<br><br> |
| - | First, the | + | First, the periplasm thickness of E. coli is as small as 10nm. In our project, we use human proteins. Almost all human proteins are much bigger than E. coli’s proteins. (They are over 10nm*10nm*10nm.) Furthermore, in our project, FGFR has 20nm height. Therefore, this protein cannot fit in periplasm, and our pathway cannot work completely.[3] <br><br> |
| - | Second, the average E. coli protein has only 360 residues, 5nm diameter, and 40kD MW. As we mentioned, our project’s proteins have over 1000residues, 10nm diameter and 50kD MW. Thus, the possibility | + | Second, the average E. coli protein has only 360 residues, 5nm diameter, and 40kD MW. As we mentioned, our project’s proteins have over 1000residues, 10nm diameter and 50kD MW. Thus, the possibility of E. coli having our pathway is very low. <br><br> |
| - | However, | + | However, yeast has big volume size from 3~4 µm to 40 µm, [4] and yeast is a eukaryotic, so it is more similar to human cell than E. coli. There is no physical problem of using human protein in yeast, and possibility of success is higher. Therefore, we use yeast instead of E. coli. |
<br> | <br> | ||
<br> | <br> | ||
| Line 72: | Line 103: | ||
<br> | <br> | ||
[[Image:pombe.png|center]] | [[Image:pombe.png|center]] | ||
| - | <span style = font-size:15px><center><b>Fig | + | <span style = font-size:15px><center><b>Fig 7. Schizosaccharomyces pombe homologus recombinant</b></center></span> |
<br> | <br> | ||
<br> | <br> | ||
| Line 84: | Line 115: | ||
[[Image:Int5.png|Biochemical Receptors|center|650px]] | [[Image:Int5.png|Biochemical Receptors|center|650px]] | ||
<br> | <br> | ||
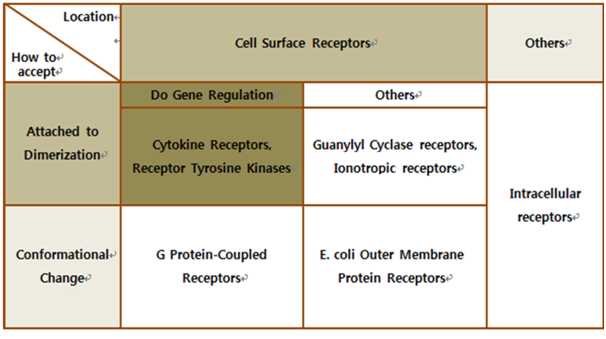
| - | The upper table represents | + | The upper table represents how we chose our biological receptor. First of all, since our genetically engineered machine has to detect the mycobacterium directly, we should select cell surface receptors. Next, we should consider using fusion antibody-receptors. We simply used gene combination (add antibody to biological receptor instead of original receipt part). If the receptor has property of changing its conformation upon acceptance of input signal, we believe that our fusion antibody receptor cannot operate well because antibodies characteristics (antigen size, antibody size, etc.) are different. They don’t have special receipt part in a whole receptor, of course, so whole receptor is important receipt part; so, we cannot create fusion-antibody-receptor with this protein. Therefore, we choose ‘Dimerization’ receptor. Most of these receptors have immunoglobulin-like receipt part, which can be alternated by antibodies easily. Also, since biosensors should activate gene expression, it has to be one of gene-regulation related receptors. <br><br> |
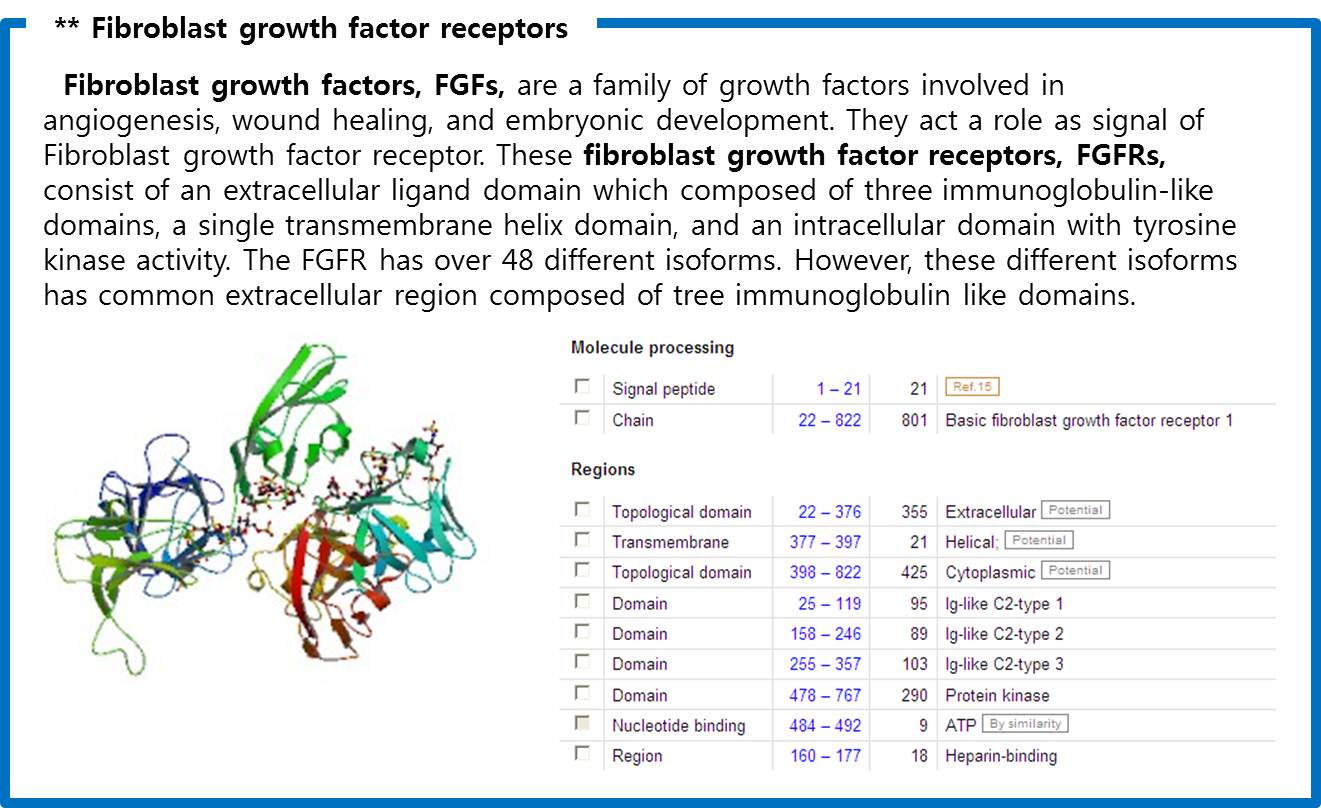
| - | Finally we choose | + | Finally we choose “cytokine receptors and receptor tyrosine kinases”. Among them <b>Fibroblast Growth Factor Receptor (FGFR)</b> in receptor tyrosine kinases was selected. |
<br> | <br> | ||
[[Image:FGFR1.jpg|center|600px]] | [[Image:FGFR1.jpg|center|600px]] | ||
| - | <span style=font-size:15px><center><b>Fig | + | <span style=font-size:15px><center><b>Fig 8. Crystal structure of a ternary FGF-FGFR-heparin complex</b></center></span> |
<br> | <br> | ||
<br> | <br> | ||
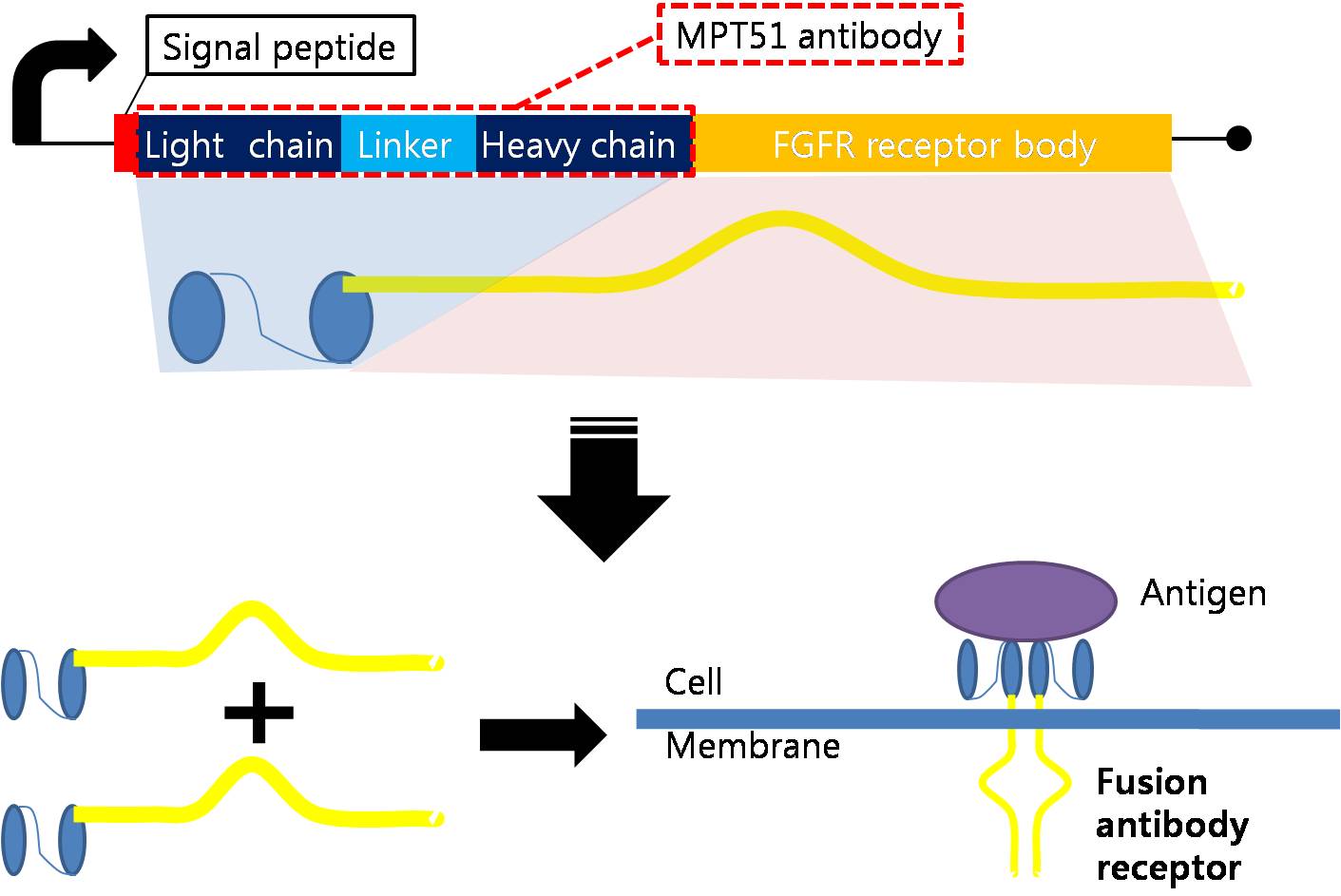
| - | === Fusion Antibody Receptor | + | === Designing Fusion Antibody Receptor === |
| - | | + | Fusion antibody-receptor is a protein whose receptor genetically combines with a specific antibody. The following figures show fusion antibody-receptor in details. |
<br> | <br> | ||
<center>[[Image:fusion antibody receptor.jpg|700px]]</center> | <center>[[Image:fusion antibody receptor.jpg|700px]]</center> | ||
<br> | <br> | ||
| - | The fusion antibody-receptor is designed by simple gene modification. We altered the | + | The fusion antibody-receptor is designed by simple gene modification. We altered the mycobacterium tuberculosis antigen detecting antibody single chain instead of the FGFR receptor’s original antigen accepting part (immunoglobulin-like parts): Light chain part of the antibody – Linker – Heavy chain part of antibody is enough. (There are some cases of successfully making fusion antibody-receptor with this method. [4]) Then, in order to express the receptor on cell surface membrane of Schizosaccharomyces pombe, we change the original signal peptide to outer membrane signal peptide of Schizosaccharomyces pombe. Surely, the receptor should satisfy our standard for biosensor. |
| Line 107: | Line 138: | ||
<td align="center" valign="center" width="150"> | <td align="center" valign="center" width="150"> | ||
</td> | </td> | ||
| - | <td style="background:#ffffff; border:2px solid #ffffff;" width=" | + | <td style="background:#ffffff; border:2px solid #ffffff;" width="800"> |
| - | <b>Schizosaccharomyces pombe Signal peptide</b> from Cell wall integrity and stress response component 1 <br> | + | <b>''Schizosaccharomyces pombe'' Signal peptide for cell surface</b> from Cell wall integrity and stress response component 1 <br> |
Protein sequence: <br> | Protein sequence: <br> | ||
| Line 123: | Line 154: | ||
=== Signaling Pathway Research === | === Signaling Pathway Research === | ||
| - | There are many signal pathways from plasma membrane receptor to the gene regulation. Here is | + | There are many signal pathways: from plasma membrane receptor to the gene regulation. Here is a famous example of these pathways, MAP kinase pathway linked with RAS pathway. In this pathway, |
# With existence of signal molecule, receptor kinases dimerize themselves and phosphorylate each other. | # With existence of signal molecule, receptor kinases dimerize themselves and phosphorylate each other. | ||
| - | # This phosphorylated receptor dimer is cognized by Grb2 and | + | # This phosphorylated receptor dimer is cognized by Grb2 and recruits SOS protein, the Guanine nucleotide Exchange Factor (GEP). |
| - | # Recruited SOS protein | + | # Recruited SOS protein substitutes as GDP of Ras protein with GTP. |
| - | # Ras protein with GTP changes its conformation and | + | # Ras protein with GTP changes its conformation and activates its own kinase activity (Ras pathway so far). |
| - | # Ras kinase | + | # Ras kinase activates the MAP kinase kinase kinase, MAP KKK(also called as Raf protein) with phosphorylation. |
| - | # MAP KKK activates the MAP kinase kinase, MAP KK(also called as Mek) with phosphorylation | + | # MAP KKK activates the MAP kinase kinase, MAP KK(also called as Mek) with phosphorylation. |
| - | # MAP KK activates the MAP kinase, MAP K(also called as Erk) with phosphorylation | + | # MAP KK activates the MAP kinase, MAP K(also called as Erk) with phosphorylation. |
# Activated MAP Kinase phosphorylates many cytosol proteins. | # Activated MAP Kinase phosphorylates many cytosol proteins. | ||
| - | # For | + | # For instance, if MAP K phosphorylates Jun TF, phosphorylated Jun goes inside of nucleus and activates the transcription to bind to DNA (MAP pathway so far). |
| - | | + | Porting this signal pathway into the Yeast is not easy because this pathway involves many new proteins, so we had to find simpler pathway. The followings are our history of the change in pathways. You can see the details 'Memo-etc page' in our wiki. |
| - | (1) Interleukin-1 pathways (8 proteins are required | + | (1) Interleukin-1 pathways (8 proteins are required)<br> |
| - | (2) Interleukin-6 JAK-STAT pathways (6 proteins are required | + | (2) Interleukin-6 JAK-STAT pathways (6 proteins are required)<br> |
| - | (3) Fc Epsilon pathways (5 proteins are required | + | (3) Fc Epsilon pathways (5 proteins are required)<br> |
| - | (4) FGFR AP-1 pathways (4 proteins are required | + | (4) FGFR AP-1 pathways (4 proteins are required)<br> |
| - | (5) FGFR STAT1 pathways (3 proteins are required | + | (5) FGFR STAT1 pathways (3 proteins are required)<br> |
| - | Finally, we selected FGF signaling pathway which is the | + | Finally, we selected FGF signaling pathway, which is the simplest. The signal transduction step of FGF signaling pathway is as followed. |
<br> | <br> | ||
<br> | <br> | ||
| Line 152: | Line 183: | ||
=== Signaling Pathway Research : Fibroblast Growth Factor Signaling Pathway === | === Signaling Pathway Research : Fibroblast Growth Factor Signaling Pathway === | ||
[[image:FGFR STAT1 pathway.jpg|center|500px]] | [[image:FGFR STAT1 pathway.jpg|center|500px]] | ||
| - | <span style=font-size:15px><center><b>Fig. | + | <span style=font-size:15px><center><b>Fig.9 FGF signaling pathway</b></center></span> |
<br> | <br> | ||
| - | | + | The figures show simple pathway of FGF (Fibroblast Growth Factor) and FGFR (Fibroblast Growth Factor).<br> FGF is a growth factor related with wound healing and embryonic development. |
<br> | <br> | ||
| Line 164: | Line 195: | ||
<td align="center" valign="center" width="500"> | <td align="center" valign="center" width="500"> | ||
[[image:syndecan2 function.jpg|center|400px]] | [[image:syndecan2 function.jpg|center|400px]] | ||
| - | <center><span style=font-size:15px><b>Fig. | + | <center><span style=font-size:15px><b>Fig.10 Unneccesary Syndecan-2 Function</b></span></center> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 171: | Line 202: | ||
<br> | <br> | ||
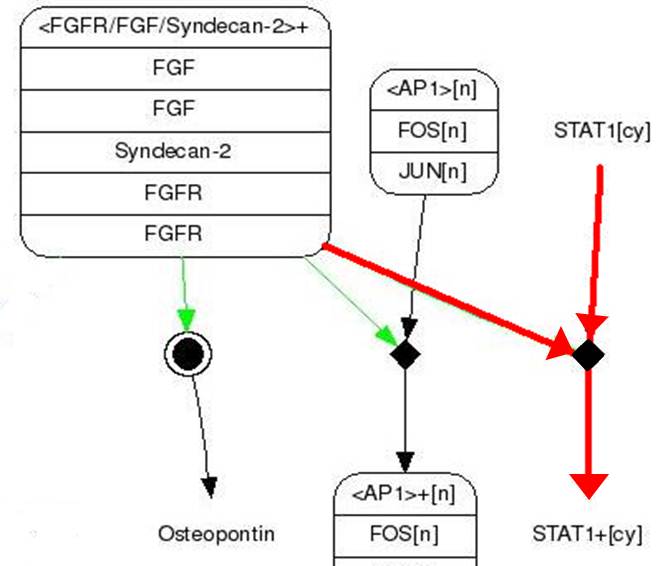
| - | # FGF binds to FGFR | + | # FGF binds to FGFR (Syndecan-2 helps in this process). |
| - | # STAT1 sticks to | + | # STAT1 sticks to phosphorylated FGFR and then it gets phosphoryl group from FGFR (Phosphorylation). |
| - | # Two | + | # Two phosphorylated STAT1s attach to each other and form a dimer. |
| - | # | + | # The dimer goes inside a nucleus and binds to GAS promoter. |
| - | # | + | # The promoted gene expresses. |
<br> | <br> | ||
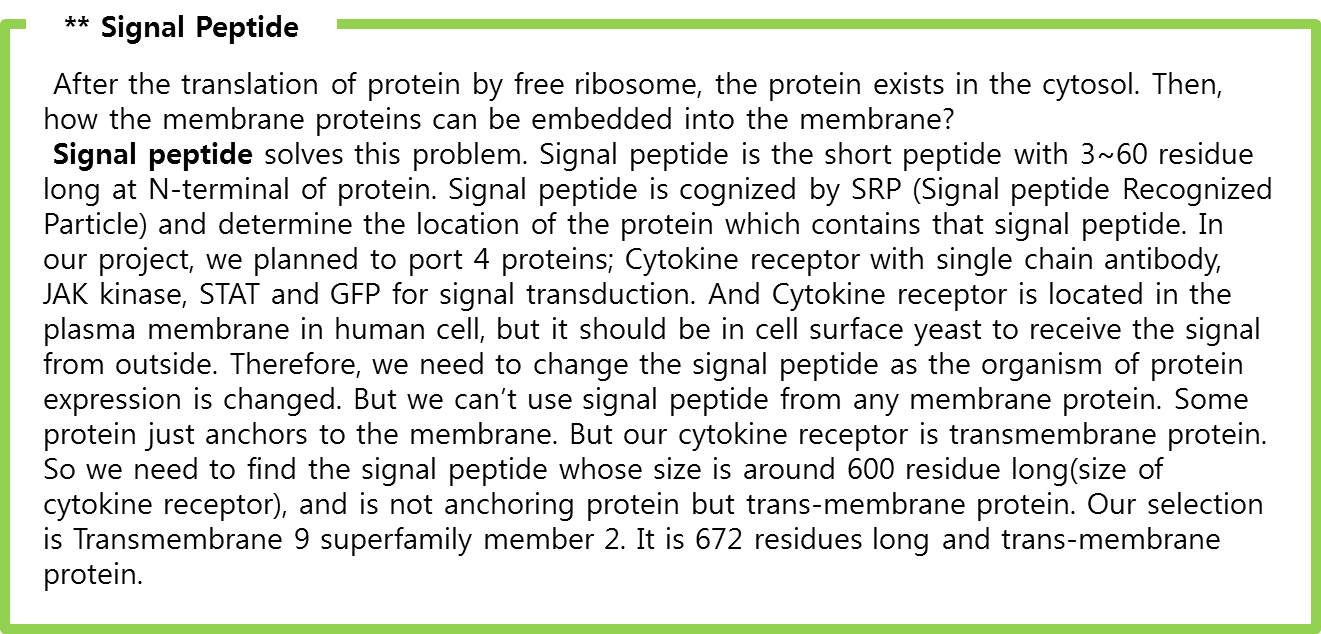
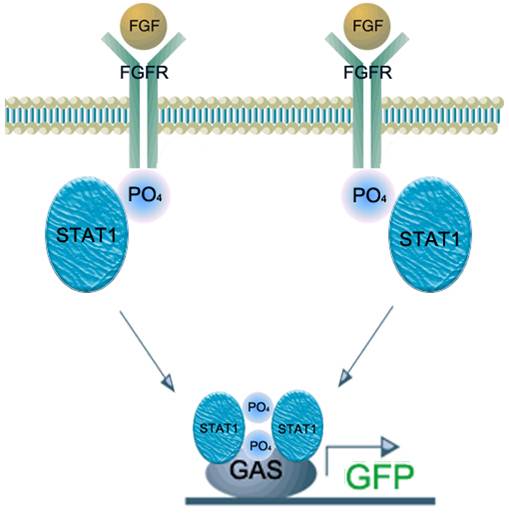
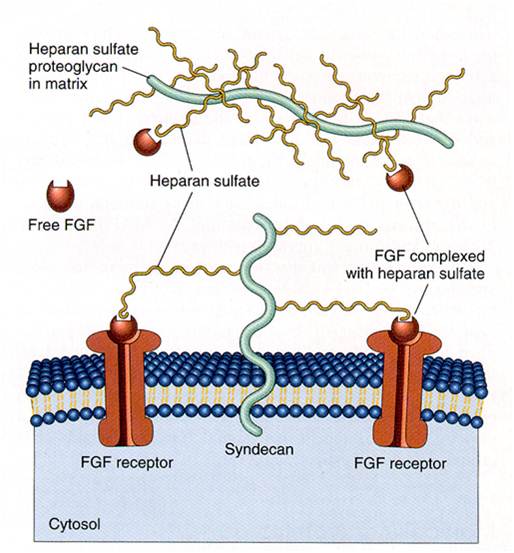
| - | We applied this pathway to our project. Actually, | + | We applied this pathway to our project. Actually, syndecan is a cell surface proteoglycan. Its core protein spans the plasma membrane and contains three heparan sulfate chains and two chondroitin sulfate chains (not shown). The proteoglycans can modulate the activity of fibroblast growth factor (FGF). Free FGF binds poorly to the FGF receptor. Binding of FGF to the heparan sulfate chains like those on syndecan allows FGF to bind efficiently its receptor. Binding of FGF to heparan sulfate in the ECM protects FGF and forms a reservoir of the growth factor. In addition we use antibody to detect tuberculosis antigen and GFP to confirm whether antigen is bound. Therefore, we don’t need Syndecan-2 because there is no FGF particle in this project. |
<br> | <br> | ||
| - | | + | In conclusion, we need three genes (Fusion FGFR, STAT1, GFP with GAS promoter) in our project. |
<br> | <br> | ||
<br><br> | <br><br> | ||
| Line 185: | Line 216: | ||
=== Design Gene Activation For Output === | === Design Gene Activation For Output === | ||
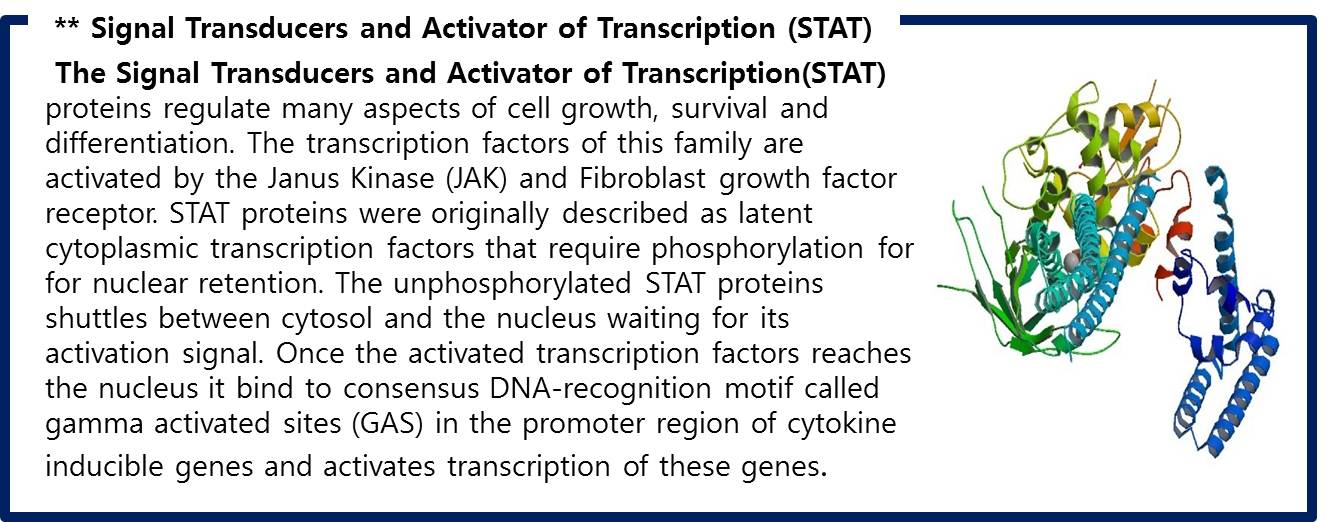
[[Image:STAT1.jpg|center|600px]] | [[Image:STAT1.jpg|center|600px]] | ||
| + | <span style=font-size:15px><center><b>Fig 11. Structural bases of unphosphorylated STAT1 association and receptor binding</b></center></span> | ||
<br> | <br> | ||
| - | STAT1 proteins with the formation of STAT1/1 dimers are confirmed using probe, namely the GAS probe. '''GAS probe'''(''G''amma ''A''ctivation ''S''ite probe) is | + | STAT1 proteins with the formation of STAT1/1 dimers are confirmed using probe, namely the GAS probe. '''GAS probe'''(''G''amma ''A''ctivation ''S''ite probe) is an acute phase responsive element of the IL gene promoter and known to bind to STAT1 dimer proteins. Using this pathway, we placed '''green fluorescent protein (GFP) gene''' instead of cytokine inducible genes. We can show GFP expression at the previous pathway. |
<br><br> | <br><br> | ||
[[Image:Int9.png|center]] | [[Image:Int9.png|center]] | ||
| + | <span style=font-size:15px><center><b>Fig 12. Proteína fluorescente verde – história e perspectivas</b></center></span> | ||
| + | <br> | ||
<br><br> | <br><br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<br> | <br> | ||
| - | : | + | <span style=font-size:30px><font face="Maiandra GD"><b>For Success! Evidences of our project’s feasibility </b></font></span> |
| - | + | ---- | |
| - | + | [[Image:success KAIST.jpg|400px|center]] | |
| - | + | ||
| - | </ | + | <span style=font-size:15px><center><b>Fig 13. Success KAIST!</b></center></span> |
| - | + | <br> | |
| - | + | <br> | |
| - | + | To successfully complete our project, we need <b>4 conditions</b>. | |
| - | </ | + | <br> |
| - | + | # FGFR should be expressed completely in yeast. | |
| - | </ | + | # FGFR and STAT1 should be located at appointed position in yeast. |
| + | # FGFR-STAT1 pathway should be operated as we expected to act normally as a transcription factor. | ||
| + | # Antibody fusion-receptor should work properly to phosphorylate sub-pathway molecules, STAT1s. | ||
| + | <br> | ||
| + | We have 4 papers that suggest our project will work. | ||
| + | <br> | ||
| + | <br> | ||
| + | <b>1.</b> [Reconstitution of Fibroblast Growth Factor Receptor Interactions in the Yeast Two Hybrid System] By Ronit A loni-Grinstein, Andrew Seddon, and Avner Yayon | ||
| + | <br> | ||
| + | >> This paper used yeast two hybrid system to show that FGFR interaction is possible in yeast. Through this paper, we knew that FGFR can be expressed completely in yeast without any functional disability and located in outer cellular membrane. | ||
| + | <br> | ||
| + | <br> | ||
| + | <b>2.</b> [Jak2-Stat5 Interactions Analyzed in Yeast (Received for publication, January 30, 1998, and in revised form, March 2, 1998)] By Fariba Barahmand-Pour, Andreas Meinke, Bernd Groner, and Thomas Decker | ||
| + | <br> | ||
| + | >> This paper proves that JAK-STAT pathway is operated completely in yeast. The JAK in yeast phosphorylates STAT5 without any trouble. FGFR belongs to tyrosine kinase receptor family, which is also JAK’s family, and STAT1 belongs to same protein family as STAT5 does. This rational suggests feasibility of our signal delivery system. Consequently, we were convinced that FGFR and STAT1 will be expressed completely to work as a signal pathway, and locate their appointed position properly. | ||
| + | <br> | ||
| + | <br> | ||
| + | <b>3.</b> [Fibroblast Growth Factor Receptor-Induced Phosphorylation of STAT1 at the Golgi Apparatus Without Translocation to the Nucleus] By LUCI´A CITORES, LING BAI,2 VIGDIS SØRENSEN, AND SJUR OLSNES2 | ||
| + | <br> | ||
| + | >> This paper shows that FGFR phosphorylates STAT1 at the golgi apparatus in human cell. The expressed FGFR at the golgi apparatus phosphorylates STAT1, and two phosphorylated STAT1 become dimerized to activate GAS promoter. The result of this study assures our FGFR-STAT1 pathway. | ||
| + | <br> | ||
| + | <br> | ||
| + | <b>4.</b> [Novel Recognition Motif on Fibroblast Growth Factor Receptor Mediates Direct Association and Activation of SNT Adapter Proteins](Received for publication, April 13, 1998) by Hong Xu, Kyung W. Lee, and Mitchell Goldfarb | ||
| + | <br> | ||
| + | >> In this paper, the researchers identified fusion FGFR, which is combined with antigen called SNT, was phosphorylated itself automatically. Through this result, we confirmed that our fusion FGFR would be phosphorylated properly when combined with the targeted antigen. | ||
| + | <br> | ||
| + | <br> | ||
| + | According to these papers and searched results, <b>we are convinced that our project’s pathway will work completely as we expected.</b> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
Latest revision as of 06:16, 17 August 2010
|
Motivation
In developing country, economic is unstable, and development in medical technology is not a priority. Unfortunately, basic medical examination like blood tests, biopsy and others need certain foundation of medical technology. In addition, some areas in developing countries do not offer electricity or access to high-tech equipments. Therefore, we need new and affordable diagnosis system especially for the diseases mentioned above. For our system, we choose TB as a target. In recent decades, diagnosis and treatment of TB have been developed. Diagnostic methods include Tuberculin Skin Test, X-ray Test for its activation and examination of the sputum. In addition, people take a blood test that involves ESR, white blood cell, CRP status checking. If necessary, CT scan and endoscopy can be performed. Most of these tests, however, require high technology and proper machines. Therefore, in developing countries, these tests are not really options. As an alternative, we introduce DiscoverY.
We suggest a system using antigen-antibody reaction. Actually, antibody cannot release perceivable signals by antigen-antibody reaction alone, so we thought of fusion antibody-receptor as the solution. Fusion antibody-receptor is a receptor which is fused with other functional protein, antibody. In this way, we can get result by gene expression which would generate GFP expression after our system is triggered by target antigens. As the result, we may develop universal sensor by changing only target antigen’s antibody in fusion antibody-receptor. It will provide relatively simple and direct way of diagnosis.
BiosensorA biosensor is a device for the detection of an analyst that combines a biological component with a physicochemical detector component.[1] In order to make biosensor with genetically engineered organism, we should consider the following basic sensing process. The ‘Receive input’ part can be alternated by represented receptor, the ‘Process’ part can be signaling pathway which changes input to output, and the ‘Extract output’ part can be something easily detectable by human. Nowadays, pigments or fluorescence proteins are usually used for output, and, in order to express these substances, we make genetically modified organism. Thus, all engineered biosensors with organisms should be modeled on (consist of) 1) biochemical receptors which can activate gene expression, 2) biological pathway, and 3) gene activation for output.
Organism Decision : Why Yeast? Why we use ‘Yeast’ instead of ‘E. coli’? E. coli control is easier than Yeast control. E. coli has faster cell cycle than Yeast. Moreover, there are many E. coli templates. But, although E. coli has many profits, we choose Yeast. Why? The following table is E. coli properties.[2]
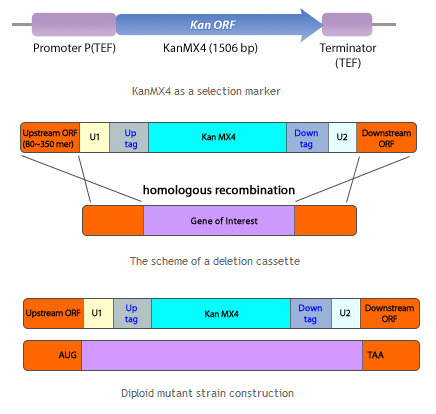
Organism Decision : Minimal genome fission yeast, Schizosaccharomyces pombe
Porting of cell signal transduction pathway is not easy for wild type organism. Most important reason is unwanted protein degradation. For wild type organism, unfamiliar protein can be harmful because it may be fragmented or misfolded proteins which disturb cellular pathway or viral proteins which may infect organisms. So wild type organism degrade unfamiliar proteins whether it is really harmful or not. And even degradation of at least one element of signal transduction pathway can disconnect whole pathway. So for successful porting of cell signal transduction pathway, inhibition of protease is important. Another problem is signal confusion. Basically, cellular signal transduction pathway is based on the protein-protein interaction(PPI). So unwanted PPI with elements of ported signal transduction pathway can confuse the signal. Of course if origin species of ported pathway is far enough from target species, we can exclude predictable unwanted PPI to avoid homologous proteins. But there are many unpredictable PPIs which can confuse the pathway. To avoid this problems, deletion of unnecessary proteins is required. And fission with minimal genome is made[5]. This yeast have only 1033 necessary genes of 5776 genes from wild type. So it may be useful to port cell signal transduction pathway from other organism(Human) without unwanted protein degradation or signal confusion.
Biochemical Receptors Selection
Finally we choose “cytokine receptors and receptor tyrosine kinases”. Among them Fibroblast Growth Factor Receptor (FGFR) in receptor tyrosine kinases was selected.
Designing Fusion Antibody Receptor Fusion antibody-receptor is a protein whose receptor genetically combines with a specific antibody. The following figures show fusion antibody-receptor in details.
Signaling Pathway ResearchThere are many signal pathways: from plasma membrane receptor to the gene regulation. Here is a famous example of these pathways, MAP kinase pathway linked with RAS pathway. In this pathway,
(1) Interleukin-1 pathways (8 proteins are required)
Signaling Pathway Research : Fibroblast Growth Factor Signaling Pathway
Design Gene Activation For Output
|
 "
"