Team:KAIST-Korea/Notebook/Memo/Etc
From 2010.igem.org
Yangstefano (Talk | contribs) |
|||
| (8 intermediate revisions not shown) | |||
| Line 9: | Line 9: | ||
<table width="100%"> | <table width="100%"> | ||
<tr> | <tr> | ||
| - | <td> | + | <td align=center> |
<html><a href="https://2010.igem.org/Team:KAIST-Korea/Notebook/Memo/Data"><img src="https://static.igem.org/mediawiki/2010/thumb/5/54/Data.jpg/230px-Data.jpg"></a></html> | <html><a href="https://2010.igem.org/Team:KAIST-Korea/Notebook/Memo/Data"><img src="https://static.igem.org/mediawiki/2010/thumb/5/54/Data.jpg/230px-Data.jpg"></a></html> | ||
<html><a href="https://2010.igem.org/Team:KAIST-Korea/Notebook/Memo/Info"><img src="https://static.igem.org/mediawiki/2010/thumb/e/e9/Info.jpg/230px-Info.jpg"></a></html> | <html><a href="https://2010.igem.org/Team:KAIST-Korea/Notebook/Memo/Info"><img src="https://static.igem.org/mediawiki/2010/thumb/e/e9/Info.jpg/230px-Info.jpg"></a></html> | ||
| Line 15: | Line 15: | ||
<html><a href="https://2010.igem.org/Team:KAIST-Korea/Notebook/Memo/Etc"><img src="https://static.igem.org/mediawiki/2010/thumb/3/39/Etc.jpg/230px-Etc.jpg"></a></html> | <html><a href="https://2010.igem.org/Team:KAIST-Korea/Notebook/Memo/Etc"><img src="https://static.igem.org/mediawiki/2010/thumb/3/39/Etc.jpg/230px-Etc.jpg"></a></html> | ||
</td></tr></table> | </td></tr></table> | ||
| + | |||
| + | <table width="100%" border="0" cellpadding="20px"> | ||
| + | <tr> | ||
| + | <td> | ||
== <span style=font-size:20px> <b> Etc. </b> </span> <span style=font-size:15px>We handle information and idea at ETC </span> == | == <span style=font-size:20px> <b> Etc. </b> </span> <span style=font-size:15px>We handle information and idea at ETC </span> == | ||
<br> | <br> | ||
| Line 58: | Line 62: | ||
So, from ATP, we can make charge gradient, to use this we can make electric energy. In this case, we don't use living cells.<br> | So, from ATP, we can make charge gradient, to use this we can make electric energy. In this case, we don't use living cells.<br> | ||
Just we need ATPase. | Just we need ATPase. | ||
| + | <br><br> | ||
| + | <span style=font-size:15px> <b> Stanford team </b> </span> | ||
| + | <br> | ||
| + | Stanford team's large protein secretion system is hard to use with liposome.<br> | ||
| + | Stanford team combined interleukin6 gene to<br> | ||
| + | :HylA signal sequence<br> | ||
| + | :HylR, HylC, IS1 : Front flanking sequence<br> | ||
| + | :HylB, HylD : Rear flanking sequence<br> | ||
| + | :tolC : channel protein<br> | ||
| + | HylA is secretion inducing sequence, and the others are induced by HylA and make channel to pump proteins to outside of the cell.<br> | ||
| + | It is different from liposome production.<br> | ||
| + | So we need to check another way for liposome based secretion. | ||
| + | <br><br> | ||
| + | <span style=font-size:15px> <b> Bacterial Secretory Vesicle </b> </span><br> | ||
| + | The appended file says that Bacterial Secretory Vesicle is hard to use. | ||
| + | <br><br> | ||
| + | <span style=font-size:15px> <b> Cancer diagnosis marker </b> </span><br> | ||
| + | I couldn’t find thesis about cancer diagnosis by bacterial method in NCBI.<br> | ||
| + | It can be both good and bad news.<br> | ||
| + | Attached file is a paper of Prof. Park published in PLoS ONE.<br> | ||
| + | It says that antibody detects surface marker of cancer tissue.<br> | ||
| + | It seems like detecting it by 4 kinds of color reaction.<br> | ||
| + | Maybe we could find another marker from another paper by prof.Lee Do Hyun, for example.<br> | ||
| + | The general concept is to transfer the system to bacteria. | ||
| + | <br><br> | ||
| + | <span style=font-size:15px> <b> BBa_K126002 </b> </span><br> | ||
| + | |||
| + | The result of blastx of 'BBa_K126002' is<br> | ||
| + | 'vascular endothelial growth factor receptor 2[Mus musculus](NCBI Reference Sequence: NP_034742.2)'.<br> | ||
| + | And the result of blast of refseq rna is<br> | ||
| + | 'Mus musculus kinase insert domain protein receptor (Kdr), mRNA (NCBI Reference Sequence: NM_010612.2)'.<br> | ||
| + | We now know that the kinase generally come from VEGFR of mouse, so we are searching that the mechanism how VEGFR accerelates growth.<br> | ||
| + | Our purpose is that we intercept the signal of VEGFR's growth accerelating mechanism to induce GFP revelation. | ||
| + | <br><br> | ||
| + | <span style=font-size:15px> <b> G-protein Signaling </b> </span><br> | ||
| + | |||
| + | There is a signaling by using G-protein, but Tyrosing kinase.<br> | ||
| + | You could find more information with this address.<br> | ||
| + | :https://2008.igem.org/Team:Illinois/Antibody_GPCR_Fusion<br> | ||
| + | We found the tyrosine receptor's identity using 'BLASTX' and 'refseq-rna DB'. | ||
<br><br> | <br><br> | ||
<span style=font-size:15px> <b> Y can D pathway </b> </span> | <span style=font-size:15px> <b> Y can D pathway </b> </span> | ||
| Line 81: | Line 125: | ||
</td></tr></table> | </td></tr></table> | ||
<br><br> | <br><br> | ||
| + | |||
| + | <table width="100%" border="0" cellpadding="20px"> | ||
| + | <tr> | ||
| + | <td> | ||
<span style=font-size:15px> <b> Why Yeast? </b> </span> | <span style=font-size:15px> <b> Why Yeast? </b> </span> | ||
<br> | <br> | ||
| Line 158: | Line 206: | ||
[[Image:Int9.png|Figure. 5|center]] | [[Image:Int9.png|Figure. 5|center]] | ||
<br><br> | <br><br> | ||
| + | |||
| + | <b>FGF signaling pathway</b> | ||
| + | <br> | ||
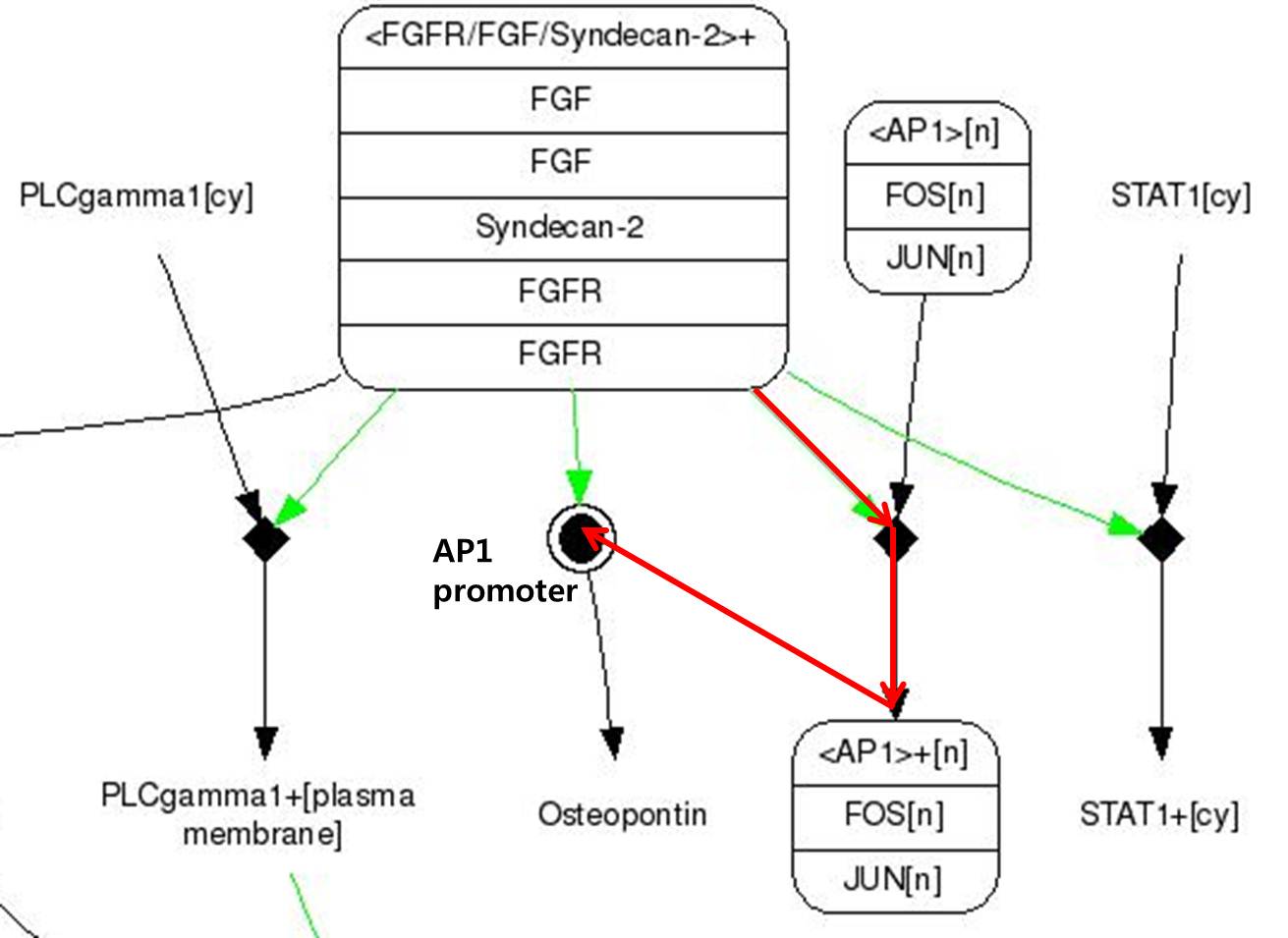
| + | We use FGF signaling pathway(FGF-2) which acts critical role in Craniosynostosis in suture closure. Whole pathway of FGF signaling change osteoblast cell activity eventually. In that one, We focused on osteopontin expression. | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | [[Image:FGF signaling pathway.jpg|center|600px]] | ||
| + | <span style = font-size:15px><center><b>Fig 8. FGF signaling Pathway. Red line pathway is selected in our project.</b></center></span> | ||
| + | <br> | ||
| + | |||
| + | # FGF-2s(Fibroblast Growth Factor-2) binds to FGFR(Fibroblast Growth Factor Receptor) | ||
| + | # There are Syndecan-2 which help signal pathway of FGF in membrane. It make FGFR to activate to pass the signal. | ||
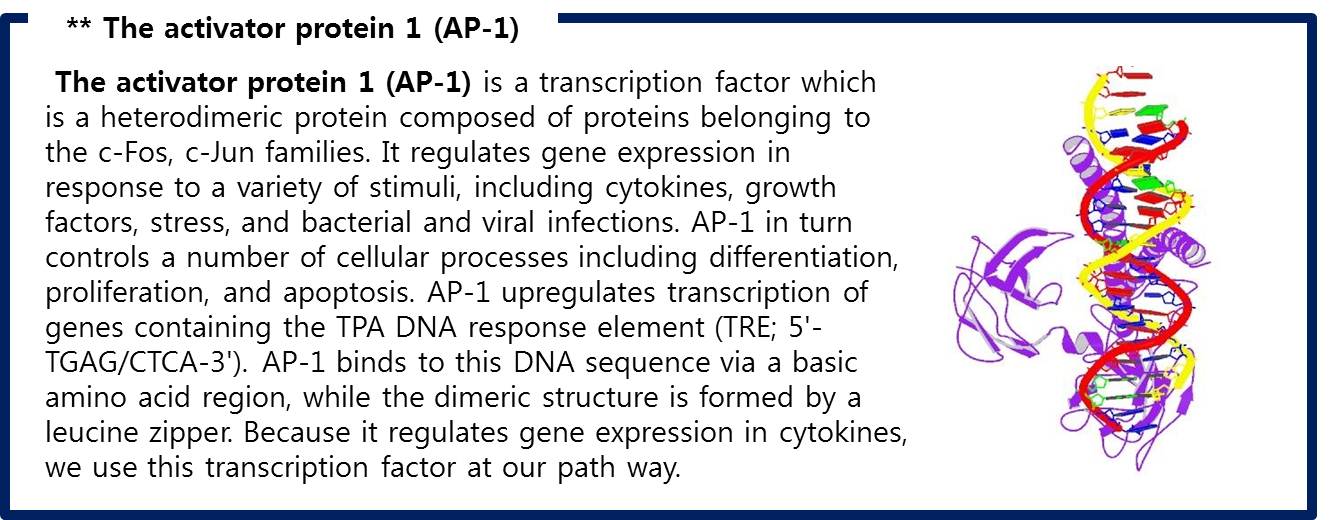
| + | # There are FOS and JUN proteins in cytoplasm.(In addtion, FOS and JUN mRNA synthesize more when FGF-2 signal binds.) Especially, c-FOS and c-JUN attach each other and form AP-1(Activate Protein 1). | ||
| + | # Changed FGFR and FGF-2 composition activate AP-1. | ||
| + | # Activated AP-1 can bound to DNA and osteopontin gene is expressd. | ||
| + | <br><br> | ||
| + | |||
| + | Like this,FGF signaling pathways have only 5 steps while RAS-MAP pathway require 9 steps, we decide to port <b>FGF signaling pathway</b> to the Yeast.<br> The introduction of <b>Syndecan-2</b> and <b>AP-1</b> are below. | ||
| + | |||
| + | |||
| + | <br> | ||
| + | [[Image:Syndecan-2.jpg|center|600px]] | ||
| + | <span style = font-size:15px><center><b>Fig 9. PDZ-like domain of syndecan-2 </b></center></span> | ||
| + | <br> | ||
| + | [[Image:AP1.jpg|center|600px]] | ||
| + | <span style = font-size:15px><center><b>Fig 10. AP-1 (c-FOS + c-JUN) bound to DNA </b></center></span> | ||
| + | <br> | ||
== referance == | == referance == | ||
| Line 170: | Line 246: | ||
::http://www.rcsb.org/pdb/explore/explore.do?structureId=1RUT | ::http://www.rcsb.org/pdb/explore/explore.do?structureId=1RUT | ||
:Fig 11. Roberto, Proteína fluorescente verde – história e perspectivas, Química de Produtos Naturais (2009) | :Fig 11. Roberto, Proteína fluorescente verde – história e perspectivas, Química de Produtos Naturais (2009) | ||
| + | :Fig 9. PDZ-like domain of syndecan-2 | ||
| + | ::RCSB Protein Database, "Solution structure of the atypical PDZ-like domain of synbindin" | ||
| + | :Fig 10. AP-1 (c-FOS + c-JUN) bound to DNA | ||
| + | ::RCSB Protein Database, "STRUCTURE OF THE DNA BINDING DOMAINS OF NFAT, FOS AND JUN BOUND TO DNA " | ||
| + | :[2] Garrett, R.H., and Grisham, C.M. Biochemistry, 2nd Edition (2002), pg. 32</br> | ||
| + | :[3] RCSB PDB(protein database), http://www.rcsb.org/ | ||
| + | :[4] Walker K, Skelton H, Smith K. (2002). accessdate=2009-11-28 "Cutaneous lesions showing giant yeast forms of Blastomyces dermatitidis". Journal of Cutaneous Pathology 29 (10): 616–18. doi:10.1034/j.1600-0560.2002.291009.x | ||
| + | :[5] Dong-Uk Kim et.al, "Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe", Nat.Biotechnol(2010) | ||
| + | :[6] Uniprot Database “Interleukin-6 receptor subunit alpha”; http://www.uniprot.org/uniprot/P08887 | ||
| + | :[7] Lichtman, Andrew H.; Abbas, Abul K. Cellular and molecular immunology (5th ed.). (2003). Philadelphia Saunders. ISBN 0-7216-0008-5. | ||
| + | :[8] SR Roffler1,6, H-E Wang2,6, H-M Yu2, W-D Chang3,4, C-M Cheng3,4, Y-L Lu5, B-M Chen1 and T-L Cheng3,4. A membrane antibody receptor for noninvasive imaging of gene expression. Gene Therapy. Nature (2005) | ||
| + | :[9] Martine I. Darville, Ye-Shih Ho and Decio L. Eizirik, NF-B Is Required for Cytokine-Induced Manganese Superoxide Dismutase Expression in Insulin-Producing Cells, The Endocrine Society(2000) | ||
| + | |||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
Latest revision as of 05:10, 17 October 2010
|
 "
"