Team:Heidelberg/Notebook/ViroBytes/October
From 2010.igem.org
(→27/09/2010) |
(→18/10/2010) |
||
| (12 intermediate revisions not shown) | |||
| Line 84: | Line 84: | ||
*~200ng of fragment DNA per reaction | *~200ng of fragment DNA per reaction | ||
*ligation time ~10min | *ligation time ~10min | ||
| + | |||
| + | ==15/10/2010== | ||
| + | |||
| + | *Fragments 1-4; 5-8 assembly. | ||
| + | **ligation time 20-25min @ 16°C | ||
| + | **60ul of beads | ||
| + | **60ul Anchor solution (Nanodrop - 196ng/ul) | ||
| + | **2x wash with 100ul w/b buffer | ||
| + | **1x wash with 100ul 1x T4 ligase buffer | ||
| + | **gentle flicking only to dissolve the bead pellet from the wall | ||
| + | |||
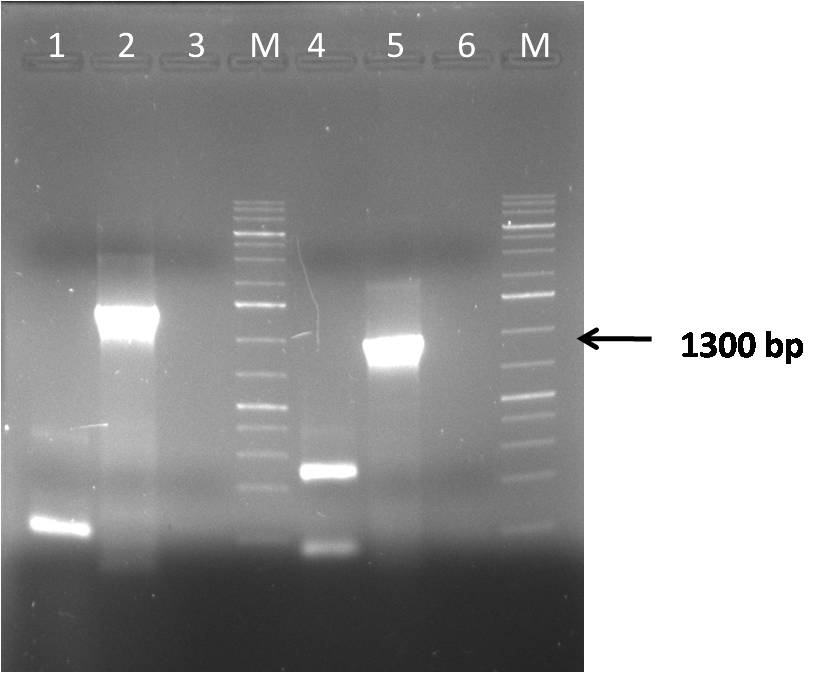
| + | [[Image:Gel_546.jpg|thumb|400px|center|]] | ||
| + | |||
| + | *1 - fragments 1-4 assembly. | ||
| + | *2 - fragments control. | ||
| + | *3 - assembly ligation mix | ||
| + | *M - DNA 1kb ladder | ||
| + | *4 - fragments 5-8 assembly. | ||
| + | *5 - fragments control. | ||
| + | *6 - assembly ligation mix | ||
| + | |||
| + | ---- | ||
| + | |||
| + | ==18/10/2010== | ||
| + | |||
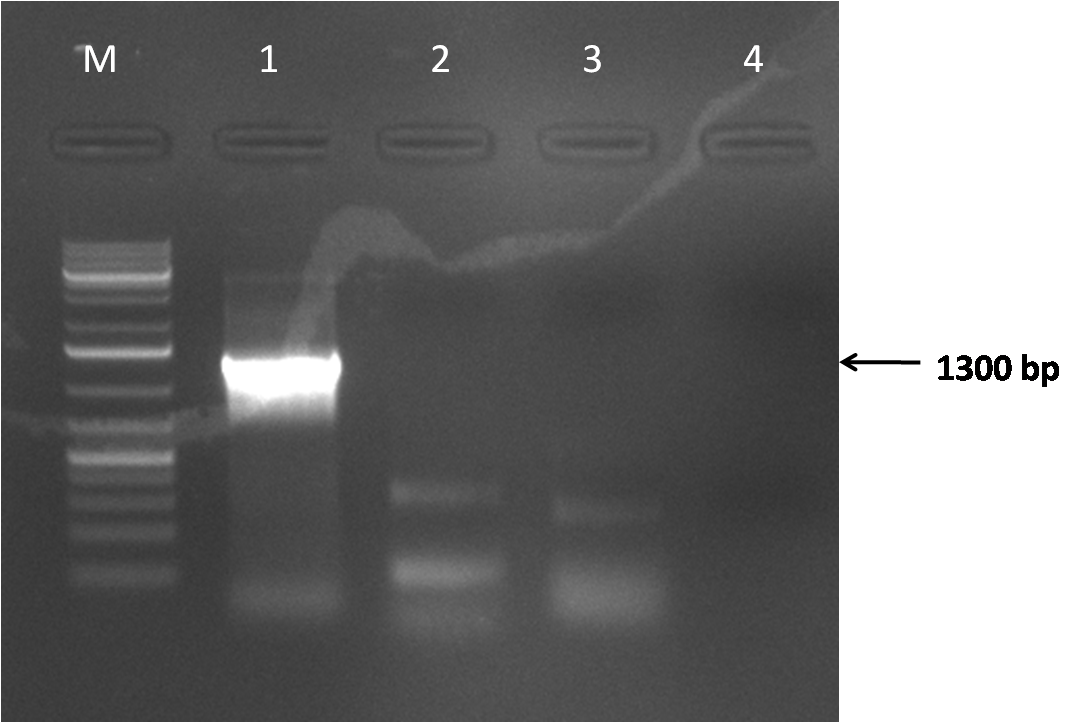
| + | *Fragments 1-4 assembly | ||
| + | |||
| + | |||
| + | Gel map: | ||
| + | |||
| + | *M - DNA 1kb ladder | ||
| + | *1 - control (fragments 1-4 from AAV1). | ||
| + | *2 - fragments 1-4 assembly | ||
| + | *3 - fragments 1-4 assembly (2nd reaction) | ||
| + | *4 - negative control | ||
| + | |||
| + | |||
| + | [[Image:Gel_547.png|thumb|500px|center|]] | ||
| + | |||
| + | ---- | ||
| + | |||
==25/10/2010== | ==25/10/2010== | ||
| Line 102: | Line 143: | ||
................................................ (1x | ................................................ (1x | ||
| - | 98 °C/15 s | + | 98 °C/15 s |
| + | |||
72 °C/30 s | 72 °C/30 s | ||
| Line 112: | Line 154: | ||
4 °C/ hold | 4 °C/ hold | ||
| + | |||
| + | |||
[[Image:hot start 25102010 b.png|thumb|350px|center|fragments 5-8 of AAV2 and 1-8 of AAV6]] | [[Image:hot start 25102010 b.png|thumb|350px|center|fragments 5-8 of AAV2 and 1-8 of AAV6]] | ||
| + | |||
| + | <br /> | ||
==26/10/2010== | ==26/10/2010== | ||
| - | * | + | *Fragment digest, Bsa1, 3hrs, purified from gel |
| + | **7ul fragment DNA | ||
| + | **3ul NEB4 10x | ||
| + | **3ul BSA 10x | ||
| + | **1ul Bsa1 | ||
| + | **16ul nuclease free water | ||
| - | + | ==27/10/2010== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ==27/ | + | |
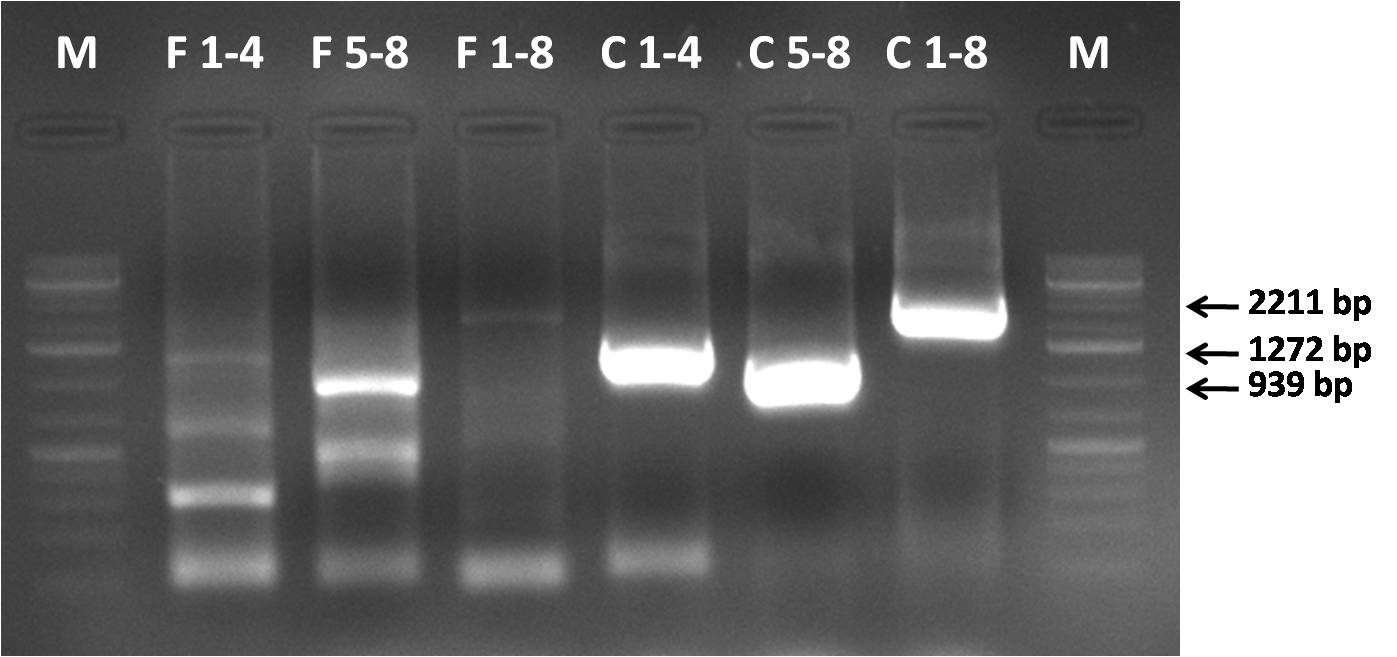
AAV 1 fragments 1-4; 5-8 and 1-8 (full capsid) assembly | AAV 1 fragments 1-4; 5-8 and 1-8 (full capsid) assembly | ||
| Line 163: | Line 196: | ||
72 °C/1 min | 72 °C/1 min | ||
| + | |||
................................................ (20x) | ................................................ (20x) | ||
Latest revision as of 03:57, 28 October 2010

October4/10/2010
5/10/2010
15/10/2010
18/10/2010
25/10/2010
PCR was set up as follows: 10 ul Phusion HF Buffer 5x 1ul of dNTP 0.5 ul of 100 um primers 2 ul of AAV template 36 ul of water ................................................ 98 °C/45 s ................................................ (1x 98 °C/15 s 72 °C/30 s ................................................ (35x) 72 °C/10 min ................................................ 4 °C/ hold
26/10/2010
27/10/2010AAV 1 fragments 1-4; 5-8 and 1-8 (full capsid) assembly Touchdown Phusion HiFi PCR was performed according to the following protocol: ................................................ 98 °C/30s ................................................ (1x 98 °C/15 s 72 °C/15 s (- 1.0 °C/cycle) 72 °C/1 min ................................................ (17x) 98 °C/15 s 55 °C/30 s 72 °C/1 min ................................................ (20x) 72 °C/10 min ................................................ (1x) 4 °C/ hold ................................................
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"