Team:Valencia/Notebook/July
From 2010.igem.org
Alejovigno (Talk | contribs) |
|||
| (36 intermediate revisions not shown) | |||
| Line 86: | Line 86: | ||
| <Font Color="#c0c0c0">28</Font> ||<Font Color="#c0c0c0">29</Font> || <Font Color="#c0c0c0">30</Font> || 1 || 2 || 3 || 4 | | <Font Color="#c0c0c0">28</Font> ||<Font Color="#c0c0c0">29</Font> || <Font Color="#c0c0c0">30</Font> || 1 || 2 || 3 || 4 | ||
|- | |- | ||
| - | | 5 || 6 || 7 || [[Team:Valencia/Notebook/July#July 8th|8]] ||[[Team:Valencia/Notebook/July#July 9th|9]] || 10 || 11 || | + | | 5 || 6 || 7 || [[Team:Valencia/Notebook/July#July 8th|8]] ||[[Team:Valencia/Notebook/July#July 9th|9]] || [[Team:Valencia/Notebook/July#July 10th|10]] || [[Team:Valencia/Notebook/July#July 11th|11]] || |
|- | |- | ||
| - | | 12 || 13 || 14 || 15 || 16 || 17 || 18 || | + | | [[Team:Valencia/Notebook/July#July 12th|12]] || [[Team:Valencia/Notebook/July#July 13th|13]] || [[Team:Valencia/Notebook/July#July 14th|14]] || [[Team:Valencia/Notebook/July#July 15th|15]] || 16 || 17 || 18 || |
|- | |- | ||
| 19 || 20 || 21 || 22 || 23 || 24 || 25 || | | 19 || 20 || 21 || 22 || 23 || 24 || 25 || | ||
| Line 171: | Line 171: | ||
<br> | <br> | ||
| - | =July | + | =July 12th= |
| + | - We made a miniprep to purify the plasmids pG1 and pLZ. | ||
| + | [[Image:miniprepGZ_12-7_3.jpg|center|thumb|200px|Dani reading the protocol and Gabi moral support for the first miniprep!!]] | ||
| + | But before you start any procedure, you have to read the protocol!!!! | ||
| - | == | + | - Then we carried out a digestion and the following electrophoresis to verify whether we had the constructions in the plasmids and to evaluate their concentration. |
| + | <gallery widths=200px heights=150px perrow=2> | ||
| + | Image:miniprepGZ_12-7_5.jpg|Gabi and the high-tech device for run the gel!! | ||
| + | Image:gel_ready_12-7_2.jpg|Gabi, Jose, Juan Angel and Dani's red hair doing the miniprep, almost done!! | ||
| + | </gallery> | ||
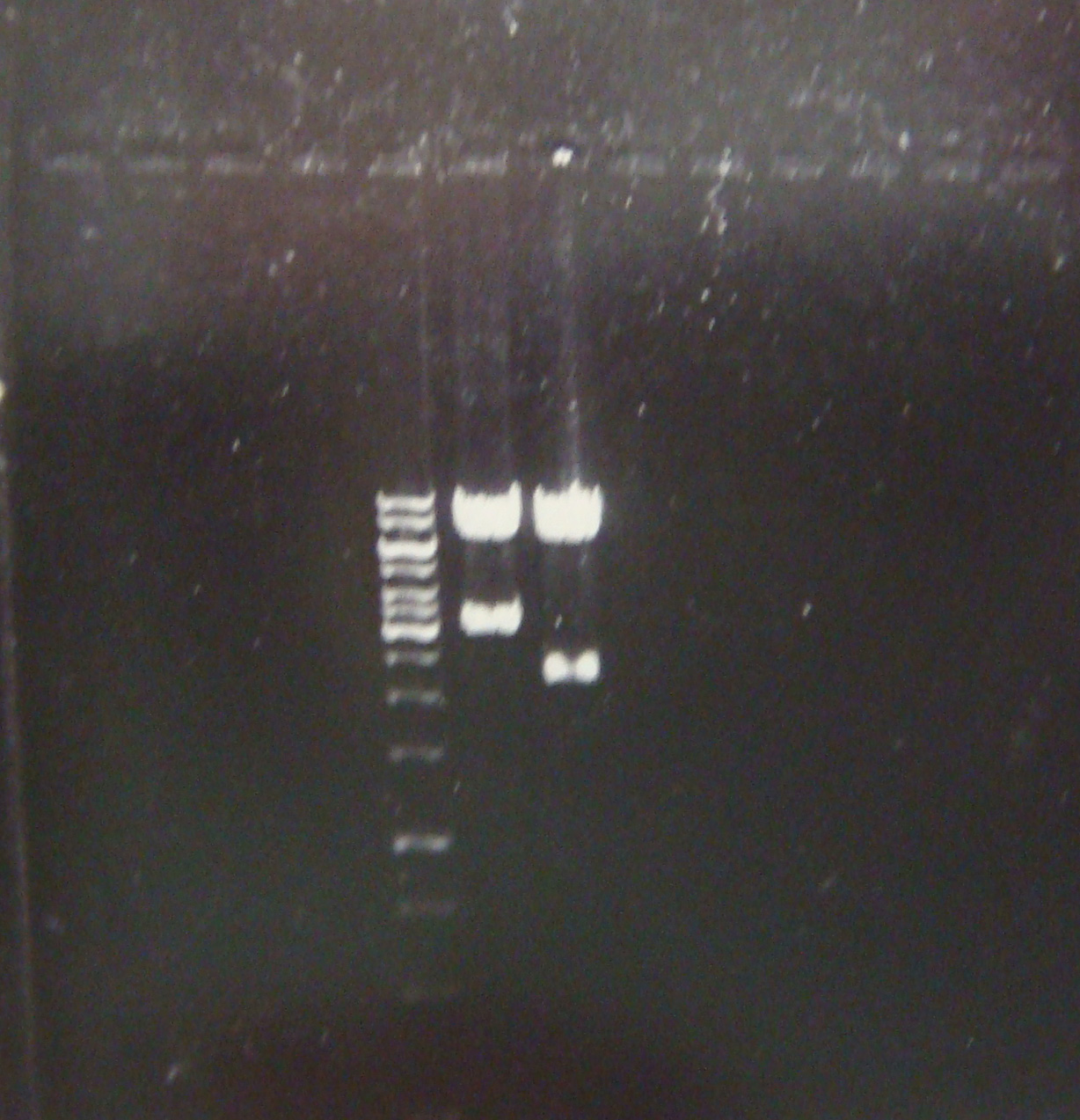
| - | + | First good result!! Here a picture of the electrophoresis. From left to right: Marker, pLZ and pG1. | |
| - | + | ||
| - | We | + | [[Image:electroforesisGZfoto_12-7_2_c.jpg|center|thumb|260px| We have the constructions and in a good concentration!]] |
| - | + | - We stored the non-digested plasmids in the refrigerator (we want to use it later on) | |
| + | - We made (Jose made) a liquid culture of ''E. coli'' with the LEA gene, and puted it into the stove at 37ºC. | ||
| + | Ale saying Jose what to do :D!! | ||
| - | == | + | <gallery widths=200px heights=200px perrow=2> |
| - | + | Image:Ecoli_liq_12-7_1.jpg|Ale saying Jose what to do :D! | |
| - | + | Image:Ecoli_liq_12-7_2.jpg|and Jose putting the liquid medium in the tube | |
| + | </gallery> | ||
| + | - We recieved a filter paper soaked with a plasmid from Kausik Li containing an ''Aplysia'' prion fused with the GR domain. Then we put the paper containing the plasmid in water in order to transform and clone it in ''E. coli'' later on. | ||
| - | =July | + | =July 13th= |
| - | We | + | - We carried out a miniprep to purify the pM2 plasmid containing the LEA gene. |
| - | + | ||
| - | + | ||
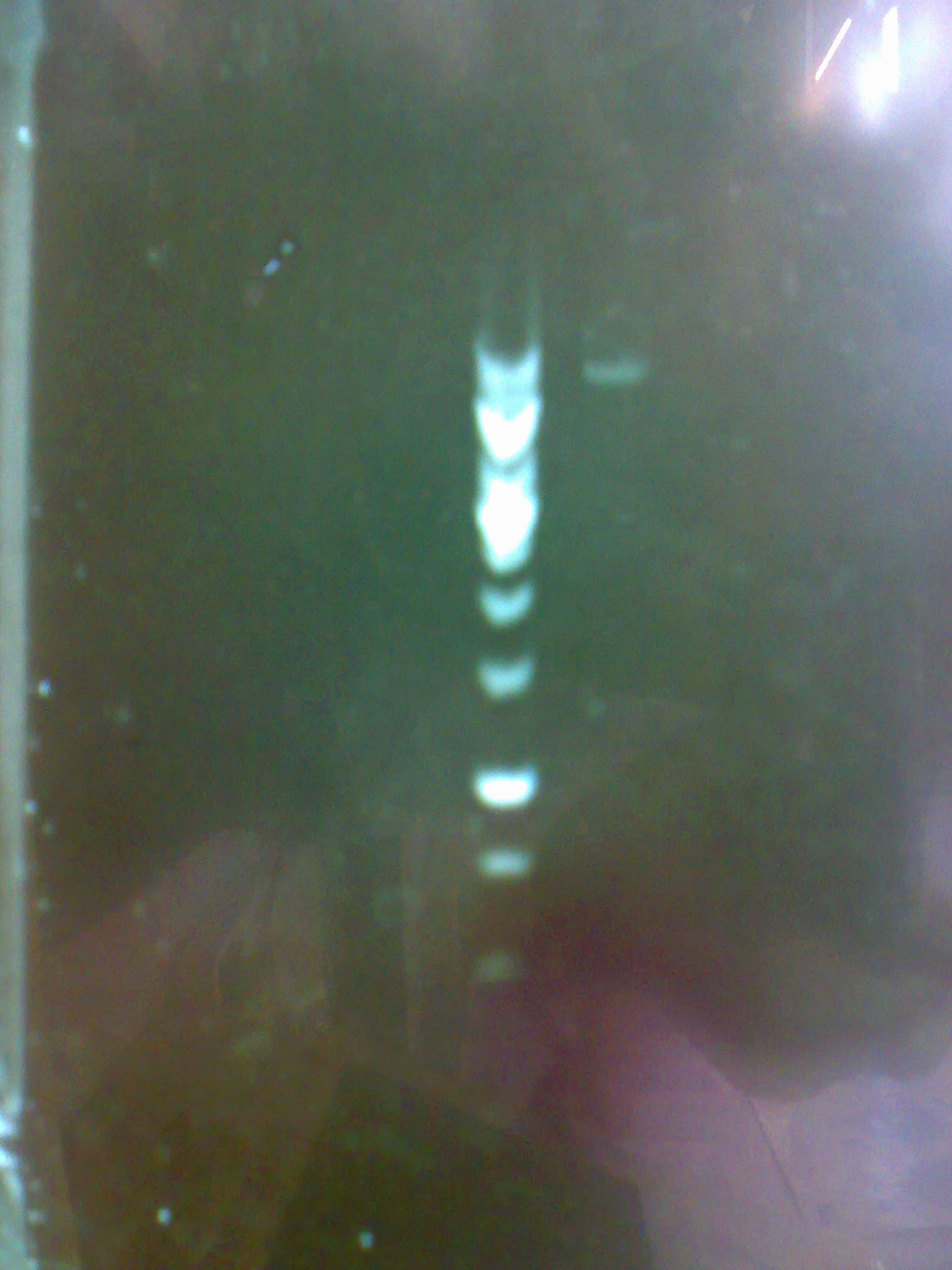
| - | + | - Then we carried out a digestion and the following electrophoresis to verify whether we had the constructions in the plasmids and to evaluate their concentration. | |
| - | + | [[Image:electropho_13-7.jpg|center|thumb|200px|Electrophoresis of the purified pM2 plasmid.]] | |
| + | - We performed a transformation of plasmid DNA containing a prionic gene into E.coli following a heat shock protocol. | ||
| + | =July 14th= | ||
| - | + | - We prepared 600ml of yeast minimum culture medium (SD) for our future yeast cultures.<br /> | |
| - | + | SD Composition: <br /> | |
| + | {| | ||
| + | |YNB w/o a.a. | ||
| + | |............. | ||
| + | |6.7 g (1.7 + 5(NH<sub>4</sub>)<sub>2</sub>SO<sub>4</sub> ) | ||
| + | |- | ||
| + | |Glucosa | ||
| + | |............. | ||
| + | |20 g | ||
| + | |- | ||
| + | |Agar | ||
| + | |............. | ||
| + | |20 g | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |............. | ||
| + | |c.s.p. 1 L | ||
| + | |} | ||
| + | : | ||
| + | - We also elaborated five amino acid and nitrogen bases solutions (Leu, His, Trp, Adenine, Uracil) for which our competent yeast strains are metabolic mutants.<br /> | ||
| + | Solutions Composition: <br /> | ||
| + | {| | ||
| + | |a.a. / b.n. | ||
| + | |............. | ||
| + | |100 mg | ||
| + | |- | ||
| + | |H<sub>2</sub>O d.d. | ||
| + | |............. | ||
| + | |50 ml | ||
| + | |} | ||
| + | : | ||
| + | We will use them in order to differentiate between plasmid transformed and non transformed yeast colonies (Only those yeast cells carrying the plasmid with the metabolic genes that our auxotrophic mutants lack will grow). | ||
| + | - No bacterial growth was observed on the dishes we planted yesterday. Therefore we re-cultured the two plasmid transformed ''E.coli'' strains (with the ''Aplysia'' and yeast prion genes) we worked on July 12th with. | ||
| - | + | - We started a culture of SC 5523 and SC 2606 yeasts strains (PSI+ and PSI-) in 5 ml of YPD. Overnight incubation at 30ºC in Emilia's Lab. | |
| - | We | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | =July | + | =July 15th= |
| - | We | + | |
| - | Then | + | - We prepared 0.5 L of YPD for stock. Four matraces of 250 ml with 50 ml each and one bottle of 0.5 L with 300 ml. |
| - | + | ||
| + | YPD Concentration | ||
| + | {| | ||
| + | |Yeast extract | ||
| + | |............. | ||
| + | |10 g | ||
| + | |- | ||
| + | |Peptone | ||
| + | |............. | ||
| + | |20 g | ||
| + | |- | ||
| + | |Dextrosa (D-glucosa) | ||
| + | |............. | ||
| + | |20 g | ||
| + | |- | ||
| + | |Agar | ||
| + | |............. | ||
| + | |10 g | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |............. | ||
| + | |c.s.p. 1 L | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | - One colony was observed in the pG1-APC dish and non in the pG1-NMG1 one. We re-planted the colony (single strip) and left it in the 37ºC stove. | ||
| + | |||
| + | - The LiAc transformation protocol for Yeasts was started | ||
| + | |||
| + | The yeast strains are going to be transformed with the pL2/GZ plasmid (GRE domain and LacZ reporter). | ||
| + | We planted tha yeasts in dishes with SD medium + Adenine, His. Also 200 µl of Try and Uracil were added in each one of the dishes, because the selection marker of the plasmid is the Leu synthesis gene. | ||
| + | |||
| + | LiAc Protocol | ||
| + | #Plant a colony in a petri dish with 5ml of YPD medium. Then overnight incubate at 30ºC. | ||
| + | #Inoculate at 0.4 OD a flask with 20 ml of YPD at ambient temperature. Inbubate at 30ºC with agitation until 1.6 OD. | ||
| + | #Pick up the cells in a 50 ml corning. Spin at 3500 rpm 3'. | ||
| + | #Discard the supernadant, and re-suspended the cell pack in a 10 ml of sterile water. Spin. | ||
| + | #Discard the water content. Re-suspended in 0.4 ml of LiAc 100mM and pass to a Eppendorf. Spin at 13000 rpm 15s. | ||
| + | #Discard LiAc. Re-suspend in 160 µl of LiAc 100mM in the vortex (Total aprox. volume 500 µl). | ||
| + | #Separate in 75 µl aliquots (each one can be used for one transformation). Spin at 13000 rpm 15s and discard LiAc. | ||
| + | #Add (important to follow the order) | ||
| + | #* 240 µl PEG 50% (p/v). | ||
| + | #* 36 µl LiAc 1 M. | ||
| + | #* 50 µl DMA carrier (2 mg/ml). | ||
| + | #* 3 µl of plasmidic DNA (with a previous bath of 5' at 100ºC, and 5' on ice). | ||
| + | #* 31 µl sterile water. | ||
| + | #Agigate in the vortex 1', until total re-suspension. | ||
| + | #Incubate at 30ºC, 30' without agitation. Then add 33µl '''something magic'''. | ||
| + | #Heat shock in 42ºC water bath for 20'. | ||
| + | #Spin at 8000 rpm for 1'. Discard supernadant. | ||
| + | #Re-suspend in 200µl of YPD and grow directly in dishes. | ||
<html> | <html> | ||
Latest revision as of 08:27, 19 July 2010
Time goes by...
(El tiempo pasa...)
Follow us:

Our main sponsors:

Our institutions:

Visitor location:
|
|
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Contents |
July 12th
- We made a miniprep to purify the plasmids pG1 and pLZ.
But before you start any procedure, you have to read the protocol!!!!
- Then we carried out a digestion and the following electrophoresis to verify whether we had the constructions in the plasmids and to evaluate their concentration.
First good result!! Here a picture of the electrophoresis. From left to right: Marker, pLZ and pG1.
- We stored the non-digested plasmids in the refrigerator (we want to use it later on)
- We made (Jose made) a liquid culture of E. coli with the LEA gene, and puted it into the stove at 37ºC. Ale saying Jose what to do :D!!
- We recieved a filter paper soaked with a plasmid from Kausik Li containing an Aplysia prion fused with the GR domain. Then we put the paper containing the plasmid in water in order to transform and clone it in E. coli later on.
July 13th
- We carried out a miniprep to purify the pM2 plasmid containing the LEA gene.
- Then we carried out a digestion and the following electrophoresis to verify whether we had the constructions in the plasmids and to evaluate their concentration.
- We performed a transformation of plasmid DNA containing a prionic gene into E.coli following a heat shock protocol.
July 14th
- We prepared 600ml of yeast minimum culture medium (SD) for our future yeast cultures.
SD Composition:
| YNB w/o a.a. | ............. | 6.7 g (1.7 + 5(NH4)2SO4 ) |
| Glucosa | ............. | 20 g |
| Agar | ............. | 20 g |
| H2O | ............. | c.s.p. 1 L |
- We also elaborated five amino acid and nitrogen bases solutions (Leu, His, Trp, Adenine, Uracil) for which our competent yeast strains are metabolic mutants.
Solutions Composition:
| a.a. / b.n. | ............. | 100 mg |
| H2O d.d. | ............. | 50 ml |
We will use them in order to differentiate between plasmid transformed and non transformed yeast colonies (Only those yeast cells carrying the plasmid with the metabolic genes that our auxotrophic mutants lack will grow).
- No bacterial growth was observed on the dishes we planted yesterday. Therefore we re-cultured the two plasmid transformed E.coli strains (with the Aplysia and yeast prion genes) we worked on July 12th with.
- We started a culture of SC 5523 and SC 2606 yeasts strains (PSI+ and PSI-) in 5 ml of YPD. Overnight incubation at 30ºC in Emilia's Lab.
July 15th
- We prepared 0.5 L of YPD for stock. Four matraces of 250 ml with 50 ml each and one bottle of 0.5 L with 300 ml.
YPD Concentration
| Yeast extract | ............. | 10 g |
| Peptone | ............. | 20 g |
| Dextrosa (D-glucosa) | ............. | 20 g |
| Agar | ............. | 10 g |
| H2O | ............. | c.s.p. 1 L |
- One colony was observed in the pG1-APC dish and non in the pG1-NMG1 one. We re-planted the colony (single strip) and left it in the 37ºC stove.
- The LiAc transformation protocol for Yeasts was started
The yeast strains are going to be transformed with the pL2/GZ plasmid (GRE domain and LacZ reporter). We planted tha yeasts in dishes with SD medium + Adenine, His. Also 200 µl of Try and Uracil were added in each one of the dishes, because the selection marker of the plasmid is the Leu synthesis gene.
LiAc Protocol
- Plant a colony in a petri dish with 5ml of YPD medium. Then overnight incubate at 30ºC.
- Inoculate at 0.4 OD a flask with 20 ml of YPD at ambient temperature. Inbubate at 30ºC with agitation until 1.6 OD.
- Pick up the cells in a 50 ml corning. Spin at 3500 rpm 3'.
- Discard the supernadant, and re-suspended the cell pack in a 10 ml of sterile water. Spin.
- Discard the water content. Re-suspended in 0.4 ml of LiAc 100mM and pass to a Eppendorf. Spin at 13000 rpm 15s.
- Discard LiAc. Re-suspend in 160 µl of LiAc 100mM in the vortex (Total aprox. volume 500 µl).
- Separate in 75 µl aliquots (each one can be used for one transformation). Spin at 13000 rpm 15s and discard LiAc.
- Add (important to follow the order)
- 240 µl PEG 50% (p/v).
- 36 µl LiAc 1 M.
- 50 µl DMA carrier (2 mg/ml).
- 3 µl of plasmidic DNA (with a previous bath of 5' at 100ºC, and 5' on ice).
- 31 µl sterile water.
- Agigate in the vortex 1', until total re-suspension.
- Incubate at 30ºC, 30' without agitation. Then add 33µl something magic.
- Heat shock in 42ºC water bath for 20'.
- Spin at 8000 rpm for 1'. Discard supernadant.
- Re-suspend in 200µl of YPD and grow directly in dishes.
 "
"